STA 326 2.0 Programming and Data Analysis with R
🛠️ Data Structures in R
Dr Thiyanga Talagala
Today's menu
Vector
Matrix
Array
Data Frame
List

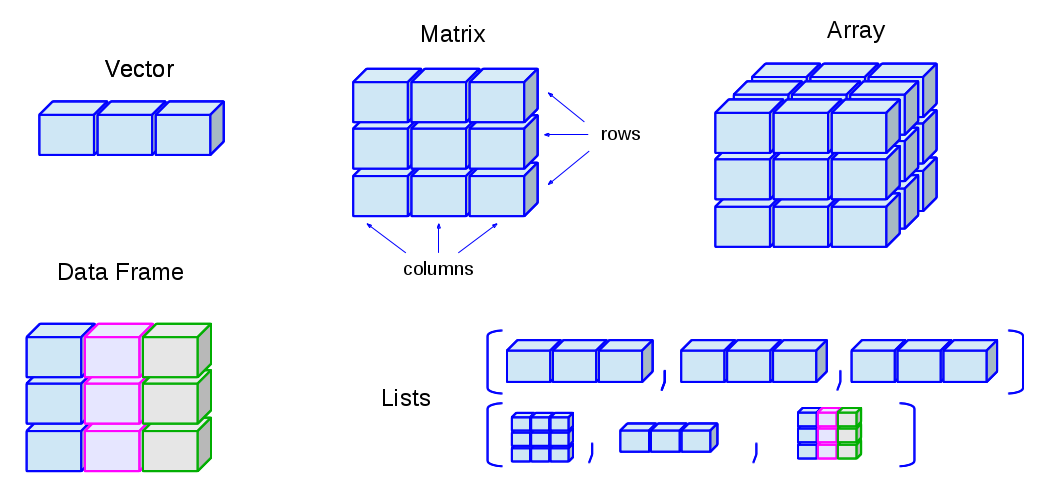
Image Credit: venus.ifca.unican.es
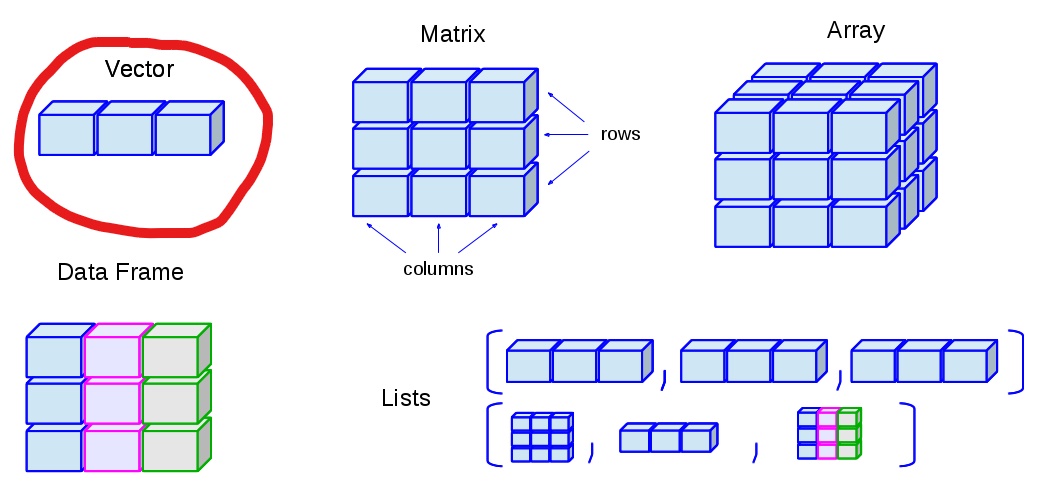
Image Credit: venus.ifca.unican.es
Recap
Write an R code to create the following vector?
[1] 1 2 3 4 5 5 4 3 2 1Recap
Write an R code to create the following vector?
[1] 1 2 3 4 5 5 4 3 2 1- Method 1
c(1, 2, 3, 4, 5, 5, 4, 3, 2, 1)Recap
Write an R code to create the following vector?
[1] 1 2 3 4 5 5 4 3 2 1- Method 1
c(1, 2, 3, 4, 5, 5, 4, 3, 2, 1)- Method 2
c(1:5, 5:1)02:00
Recap
- Name elements
a <- c(1:5, 5:1)a [1] 1 2 3 4 5 5 4 3 2 1names(a) <- c("a1", "a2", "a3", "a4", "a5", "b1", "b2", "b3", "b4", "b5")aa1 a2 a3 a4 a5 b1 b2 b3 b4 b5 1 2 3 4 5 5 4 3 2 1Operations between vectors
aa1 a2 a3 a4 a5 b1 b2 b3 b4 b5 1 2 3 4 5 5 4 3 2 1a * c(10, 100) a1 a2 a3 a4 a5 b1 b2 b3 b4 b5 10 200 30 400 50 500 40 300 20 100Vectors: subsetting
Select some particular elements (i.e., a subset) from a vector.
Vectors: Subsetting
myvec <- 1:20; myvec [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20Vectors: Subsetting
myvec <- 1:20; myvec [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20myvec[1][1] 1Vectors: Subsetting
myvec <- 1:20; myvec [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20myvec[1][1] 1myvec[5:10][1] 5 6 7 8 9 10Vectors: Subsetting (cont.)
Vectors: Subsetting (cont.)
myvec[-1] [1] 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20Vectors: Subsetting (cont.)
myvec[-1] [1] 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20myvec[myvec > 3] [1] 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20vector subsetting - special cases
myvec[0]integer(0)myvec[] [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20myvec [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20- Extract elements present in vector
afrommyvec(cont.).
a; myveca1 a2 a3 a4 a5 b1 b2 b3 b4 b5 1 2 3 4 5 5 4 3 2 1 [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20myvec %in% a [1] TRUE TRUE TRUE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE[13] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSEmyvec[myvec %in% a][1] 1 2 3 4 5Vectors: Subsetting (cont.)
myvec [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20b <- 100:105b[1] 100 101 102 103 104 105myvec[myvec %in% b]integer(0)Your turn
Generate a sequence using the code
seq(from=1, to=10, by=1).What other ways can you generate the same sequence?
Using the function
rep, create the below sequence 1, 2, 3, 4, 1, 2, 3, 4, 1, 2, 3, 4
03:00
Changing values of a vector
covid <- c(100, 30, 40, 50, -1, 100)covid[1] 100 30 40 50 -1 100covid[1] <- 50000covid[1] 50000 30 40 50 -1 100Changing values of a vector
covid <- c(100, 30, 40, 50, -1, 100)covid[1] 100 30 40 50 -1 100covid[1] <- 50000covid[1] 50000 30 40 50 -1 100covid[covid < 0] <- 0covid[1] 50000 30 40 50 0 100covid[c(1, 2)] <- c(1000, 10000)covid[1] 1000 10000 40 50 0 1002. Matrices
Matrix is a 2-dimentional and a homogeneous data structure
Syntax to create a matrix
matrix_name <- matrix(vector_of_elements, nrow=number_of_rows, ncol=number_of_columns, byrow=logical_value, # If byrow=TRUE, then the matrix is filled in by row. dimnames=list(rnames, cnames)) # To assign row names and columnsExample
matrix(1:6, nrow=2, ncol=3) [,1] [,2] [,3][1,] 1 3 5[2,] 2 4 6cont.
matrix(1:6, nrow=2) [,1] [,2] [,3][1,] 1 3 5[2,] 2 4 6matrix(1:6, ncol=3) [,1] [,2] [,3][1,] 1 3 5[2,] 2 4 6Matrix fill by rows/ columns
values <- c(10, 20, 30, 40)matrix1 <- matrix(values, nrow=2) # Matrix filled by columns (default option)matrix1 [,1] [,2][1,] 10 30[2,] 20 40matrix2 <- matrix(values, nrow=2, byrow=TRUE) # Matrix filled by rowsmatrix2 [,1] [,2][1,] 10 20[2,] 30 40Matrix fill by rows/ columns (cont.)
byrow=TRUE: matrix is filled in by row
byrow=FALSE: matrix is filled in by column
Default is by column
Naming matrix rows and columns
rnames <- c("R1", "R2")cnames <- c("C1", "C2")matrix_with_names <- matrix(values, nrow=2, dimnames=list(rnames, cnames))matrix_with_names C1 C2R1 10 30R2 20 40class function on vectors vs matrices
with vectors
vec <- 1:10class(vec)[1] "integer"with matrices
mat <- matrix(1:10, ncol=5)class(mat)[1] "matrix" "array"Matrix: subsetting
Matrix: subsetting
matrix1 [,1] [,2][1,] 10 30[2,] 20 40matraix_name[i, ] gives the ith row of a matrix
matrix1[1, ][1] 10 30matraix_name[, j] gives the jth column of a matrix
matrix1[, 2][1] 30 40Matrix: subsetting (cont.)
matraix_name[i, j] gives the ith row and jth column element
matrix1 [,1] [,2][1,] 10 30[2,] 20 40matrix1[1, 2][1] 30matrix1[1, c(1, 2)][1] 10 30Beware!
amat <- matrix(10:90, 3, 3); amat [,1] [,2] [,3][1,] 10 13 16[2,] 11 14 17[3,] 12 15 18bmat <- amat[1:2,]; bmat [,1] [,2] [,3][1,] 10 13 16[2,] 11 14 17class(bmat)[1] "matrix" "array"Beware!
amat <- matrix(10:90, 3, 3); amat [,1] [,2] [,3][1,] 10 13 16[2,] 11 14 17[3,] 12 15 18bmat <- amat[1:2,]; bmat [,1] [,2] [,3][1,] 10 13 16[2,] 11 14 17class(bmat)[1] "matrix" "array"cmat <- amat[1,]; cmat[1] 10 13 16class(cmat)[1] "integer"dmat <- amat[, 1]; dmat[1] 10 11 12class(dmat)[1] "integer"drop = FALSE
cmat <- amat[1,]; cmat[1] 10 13 16class(cmat)[1] "integer"cmat <- amat[1, , drop = FALSE]; cmat [,1] [,2] [,3][1,] 10 13 16class(cmat)[1] "matrix" "array"Diagonal elements
a <- matrix(1:9, ncol=3)a [,1] [,2] [,3][1,] 1 4 7[2,] 2 5 8[3,] 3 6 9diag(a)[1] 1 5 9Replacing elements in a matrix
a <- matrix(1:9, ncol=3)a [,1] [,2] [,3][1,] 1 4 7[2,] 2 5 8[3,] 3 6 9a[1, 1] <- 100a [,1] [,2] [,3][1,] 100 4 7[2,] 2 5 8[3,] 3 6 9diag(a) <- 0a [,1] [,2] [,3][1,] 0 4 7[2,] 2 0 8[3,] 3 6 0cbind and rbind
Matrices can be created by column-binding and row-binding with cbind() and rbind()
x <- 1:3y <- c(10, 100, 1000)cbind(x, y) # binds matrices horizontally x y[1,] 1 10[2,] 2 100[3,] 3 1000rbind(x, y) #binds matrices vertically [,1] [,2] [,3]x 1 2 3y 10 100 1000cbind and rbind (cont.)
a <- matrix(1:3, ncol=3)a [,1] [,2] [,3][1,] 1 2 3b <- matrix(c(10, 100, 1000), ncol=3)b [,1] [,2] [,3][1,] 10 100 1000cbind(a, b) # binds matrices horizontally [,1] [,2] [,3] [,4] [,5] [,6][1,] 1 2 3 10 100 1000rbind(a, b) #binds matrices vertically [,1] [,2] [,3][1,] 1 2 3[2,] 10 100 1000Matrix operations
Matrix operations
Transpose
t(x) [,1] [,2] [,3][1,] 1 2 3Matrix multiplication
y <- matrix(seq(10, 60, by=10), nrow=3)z <- x %*% yz [,1] [,2][1,] 140 320Matrix operations (cont.)
- matrix inverse is solve()
Find x in: m*x=n
solve(m, n)Your turn
05:00
ˆβ=(XTX)−1XTY
05:00
Logical operators with matrices
a <- matrix(c(1:12), nrow = 3, ncol = 4) a > 10 [,1] [,2] [,3] [,4][1,] FALSE FALSE FALSE FALSE[2,] FALSE FALSE FALSE TRUE[3,] FALSE FALSE FALSE TRUEb <- matrix(1:4, ncol=4)a > b # Error%in% operator
b %in% a[1] TRUE TRUE TRUE TRUEa %in% b [1] TRUE TRUE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSEclass(a %in% b)[1] "logical"3. Arrays
Array
data structures for storing higher dimensional data.
a homogeneous data structure.
a special case of the array is the matrix.
array(vector, dimensions, dimnames) #dimnames-as a lista <- array(c(10, 20, 30, 40, 50, 60), c(1, 2, 3))a <- array(c(10, 20, 30, 40, 50, 60), c(1, 2, 3))a, , 1 [,1] [,2][1,] 10 20, , 2 [,1] [,2][1,] 30 40, , 3 [,1] [,2][1,] 50 60Subsetting arrays
a, , 1 [,1] [,2][1,] 10 20, , 2 [,1] [,2][1,] 30 40, , 3 [,1] [,2][1,] 50 60a[1, 2, 3][1] 60a[, , 1] # Extract first entry[1] 10 20Subsetting arrays (cont.)
a, , 1 [,1] [,2][1,] 10 20, , 2 [,1] [,2][1,] 30 40, , 3 [,1] [,2][1,] 50 60a[1, ,] # All rows in each entry [,1] [,2] [,3][1,] 10 30 50[2,] 20 40 60Your turn
- Create the following matrix using the
arrayfunction
matrix(1:20, ncol=5) [,1] [,2] [,3] [,4] [,5][1,] 1 5 9 13 17[2,] 2 6 10 14 18[3,] 3 7 11 15 19[4,] 4 8 12 16 2002:00
Array with dimnames
dim1 <- c("A1", "A2"); dim2 <- c("B1", "B2", "B3"); dim3 <- c("c1", "c2", "c3", "c4")z <- array(1:24, c(2, 3, 4), dimnames = list(dim1, dim2, dim3))z, , c1 B1 B2 B3A1 1 3 5A2 2 4 6, , c2 B1 B2 B3A1 7 9 11A2 8 10 12, , c3 B1 B2 B3A1 13 15 17A2 14 16 18, , c4 B1 B2 B3A1 19 21 23A2 20 22 24dim1 <- c("A1", "A2"); dim2 <- c("B1", "B2", "B3"); dim3 <- c("c1", "c2", "c3", "c4")z <- array(1:24, c(2, 3, 4), dimnames = list(dim1, dim2, dim3))z## , , c1## ## B1 B2 B3## A1 1 3 5## A2 2 4 6## ## , , c2## ## B1 B2 B3## A1 7 9 11## A2 8 10 12## ## , , c3## ## B1 B2 B3## A1 13 15 17## A2 14 16 18## ## , , c4## ## B1 B2 B3## A1 19 21 23## A2 20 22 24
4. Data Frames
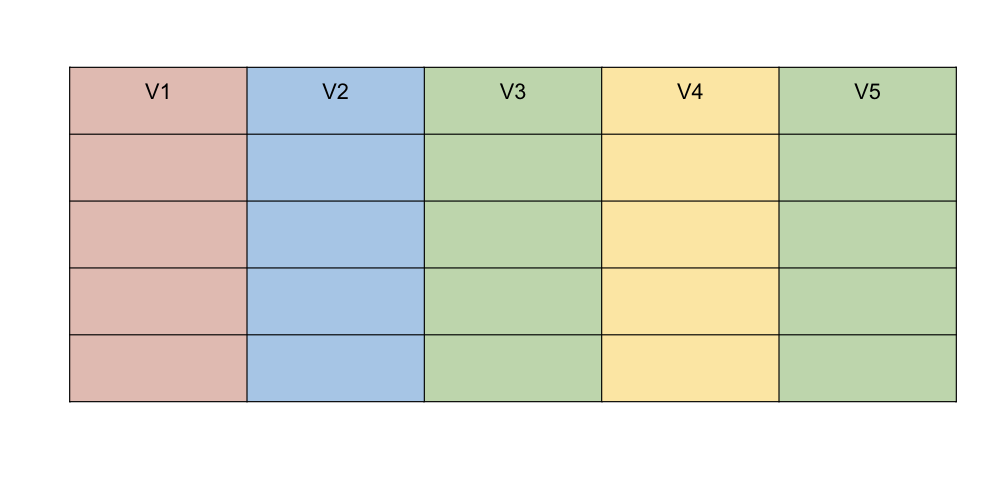
Data frames
Rectangular arrangement of data with rows corresponding to observational units and columns corresponding to variables.
More general than a matrix in that different columns can contain different modes of data.
It’s similar to the datasets you’d typically see in SPSS and MINITAB.
Data frames are the most common data structure you’ll deal with in R.

Image Credit: Hadley Wickham
Create a data frame
Syntax
name_of_the_dataframe <- data.frame( var1_name=vector of values of the first variable, var2_names=vector of values of the second variable)Example
corona <- data.frame(ID=c("C001", "C002", "C003", "C004"), Location=c("Beijing", "Wuhan", "Shanghai", "Beijing"), Test_Results=c(FALSE, TRUE, FALSE, FALSE))corona ID Location Test_Results1 C001 Beijing FALSE2 C002 Wuhan TRUE3 C003 Shanghai FALSE4 C004 Beijing FALSETo check if it is a dataframe
is.data.frame(corona)[1] TRUESome useful functions with dataframes
colnames(corona)[1] "ID" "Location" "Test_Results"length(corona)[1] 3dim(corona)[1] 4 3nrow(corona)[1] 4ncol(corona)[1] 3Some useful functions with dataframes (cont.)
summary(corona) ID Location Test_Results Length:4 Length:4 Mode :logical Class :character Class :character FALSE:3 Mode :character Mode :character TRUE :1str(corona)'data.frame': 4 obs. of 3 variables: $ ID : chr "C001" "C002" "C003" "C004" $ Location : chr "Beijing" "Wuhan" "Shanghai" "Beijing" $ Test_Results: logi FALSE TRUE FALSE FALSEConvert a matrix to a dataframe
mat <- matrix(1:16, ncol=4)mat [,1] [,2] [,3] [,4][1,] 1 5 9 13[2,] 2 6 10 14[3,] 3 7 11 15[4,] 4 8 12 16mat_df <- as.data.frame(mat)mat_df V1 V2 V3 V41 1 5 9 132 2 6 10 143 3 7 11 154 4 8 12 16Subsetting data frames
Select rows
head(mat_df) # default it shows 6 rows V1 V2 V3 V41 1 5 9 132 2 6 10 143 3 7 11 154 4 8 12 16head(mat_df, 3) # To extract only the first three rows V1 V2 V3 V41 1 5 9 132 2 6 10 143 3 7 11 15tail(mat_df, 2) V1 V2 V3 V43 3 7 11 154 4 8 12 16Subsetting data frames
mat_df V1 V2 V3 V41 1 5 9 132 2 6 10 143 3 7 11 154 4 8 12 16To select some specific rows
mat_df[4, ] V1 V2 V3 V44 4 8 12 16index <- c(1, 3)mat_df[index, ] V1 V2 V3 V41 1 5 9 133 3 7 11 15Subsetting data frames: select columns
mat_df V1 V2 V3 V41 1 5 9 132 2 6 10 143 3 7 11 154 4 8 12 16Select column(s) by variable names
mat_df$V1 # Method 1[1] 1 2 3 4mat_df[, "V1"] # Method 2[1] 1 2 3 4Subsetting data frames: select columns
mat_df V1 V2 V3 V41 1 5 9 132 2 6 10 143 3 7 11 154 4 8 12 16Select column(s) by index
mat_df[, 2][1] 5 6 7 8Your turn
03:00
Built-in dataframes
Built-in dataframes
data(iris)Use help function to find more about the iris dataset.
iris dataset
head(iris) Sepal.Length Sepal.Width Petal.Length Petal.Width Species1 5.1 3.5 1.4 0.2 setosa2 4.9 3.0 1.4 0.2 setosa3 4.7 3.2 1.3 0.2 setosa4 4.6 3.1 1.5 0.2 setosa5 5.0 3.6 1.4 0.2 setosa6 5.4 3.9 1.7 0.4 setosa
head(iris) Sepal.Length Sepal.Width Petal.Length Petal.Width Species1 5.1 3.5 1.4 0.2 setosa2 4.9 3.0 1.4 0.2 setosa3 4.7 3.2 1.3 0.2 setosa4 4.6 3.1 1.5 0.2 setosa5 5.0 3.6 1.4 0.2 setosa6 5.4 3.9 1.7 0.4 setosa
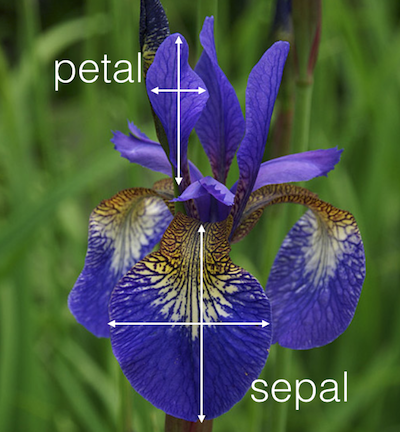
head(iris) Sepal.Length Sepal.Width Petal.Length Petal.Width Species1 5.1 3.5 1.4 0.2 setosa2 4.9 3.0 1.4 0.2 setosa3 4.7 3.2 1.3 0.2 setosa4 4.6 3.1 1.5 0.2 setosa5 5.0 3.6 1.4 0.2 setosa6 5.4 3.9 1.7 0.4 setosa- Introduced by Sir Ronald A. Fisher, 1936

Use the R dataset “iris” to answer the following questions:
How many rows and columns does iris have?
Select the first 4 rows.
Select the last 6 rows.
Select rows 10 to 20, with all columns in the iris dataset.
Select rows 10 to 20 with only the Species, Petal.Width and Petal.Length.
05:00
cont.
Create a single vector (a new object) called ‘width’ that is the Sepal.Width column of iris.
What are the column names and data types of the different columns in iris?
How many rows in the iris dataset have
Petal.Lengthlarger than 5 andSepal.Widthsmaller than 3?
05:00
5. List
List
- Lists are heterogeneous
Syntax
list_name <- list(entry1, entry2, entry3, ...)Example
first_list <-list(1:3, matrix(1:6, nrow=2), iris)List (str)
first_list <-list(1:3, matrix(1:6, nrow=2), iris)first_list[[1]][1] 1 2 3[[2]] [,1] [,2] [,3][1,] 1 3 5[2,] 2 4 6[[3]] Sepal.Length Sepal.Width Petal.Length Petal.Width Species1 5.1 3.5 1.4 0.2 setosa2 4.9 3.0 1.4 0.2 setosa3 4.7 3.2 1.3 0.2 setosa4 4.6 3.1 1.5 0.2 setosa5 5.0 3.6 1.4 0.2 setosa6 5.4 3.9 1.7 0.4 setosa7 4.6 3.4 1.4 0.3 setosa8 5.0 3.4 1.5 0.2 setosa9 4.4 2.9 1.4 0.2 setosa10 4.9 3.1 1.5 0.1 setosa11 5.4 3.7 1.5 0.2 setosa12 4.8 3.4 1.6 0.2 setosa13 4.8 3.0 1.4 0.1 setosa14 4.3 3.0 1.1 0.1 setosa15 5.8 4.0 1.2 0.2 setosa16 5.7 4.4 1.5 0.4 setosa17 5.4 3.9 1.3 0.4 setosa18 5.1 3.5 1.4 0.3 setosa19 5.7 3.8 1.7 0.3 setosa20 5.1 3.8 1.5 0.3 setosa21 5.4 3.4 1.7 0.2 setosa22 5.1 3.7 1.5 0.4 setosa23 4.6 3.6 1.0 0.2 setosa24 5.1 3.3 1.7 0.5 setosa25 4.8 3.4 1.9 0.2 setosa26 5.0 3.0 1.6 0.2 setosa27 5.0 3.4 1.6 0.4 setosa28 5.2 3.5 1.5 0.2 setosa29 5.2 3.4 1.4 0.2 setosa30 4.7 3.2 1.6 0.2 setosa31 4.8 3.1 1.6 0.2 setosa32 5.4 3.4 1.5 0.4 setosa33 5.2 4.1 1.5 0.1 setosa34 5.5 4.2 1.4 0.2 setosa35 4.9 3.1 1.5 0.2 setosa36 5.0 3.2 1.2 0.2 setosa37 5.5 3.5 1.3 0.2 setosa38 4.9 3.6 1.4 0.1 setosa39 4.4 3.0 1.3 0.2 setosa40 5.1 3.4 1.5 0.2 setosa41 5.0 3.5 1.3 0.3 setosa42 4.5 2.3 1.3 0.3 setosa43 4.4 3.2 1.3 0.2 setosa44 5.0 3.5 1.6 0.6 setosa45 5.1 3.8 1.9 0.4 setosa46 4.8 3.0 1.4 0.3 setosa47 5.1 3.8 1.6 0.2 setosa48 4.6 3.2 1.4 0.2 setosa49 5.3 3.7 1.5 0.2 setosa50 5.0 3.3 1.4 0.2 setosa51 7.0 3.2 4.7 1.4 versicolor52 6.4 3.2 4.5 1.5 versicolor53 6.9 3.1 4.9 1.5 versicolor54 5.5 2.3 4.0 1.3 versicolor55 6.5 2.8 4.6 1.5 versicolor56 5.7 2.8 4.5 1.3 versicolor57 6.3 3.3 4.7 1.6 versicolor58 4.9 2.4 3.3 1.0 versicolor59 6.6 2.9 4.6 1.3 versicolor60 5.2 2.7 3.9 1.4 versicolor61 5.0 2.0 3.5 1.0 versicolor62 5.9 3.0 4.2 1.5 versicolor63 6.0 2.2 4.0 1.0 versicolor64 6.1 2.9 4.7 1.4 versicolor65 5.6 2.9 3.6 1.3 versicolor66 6.7 3.1 4.4 1.4 versicolor67 5.6 3.0 4.5 1.5 versicolor68 5.8 2.7 4.1 1.0 versicolor69 6.2 2.2 4.5 1.5 versicolor70 5.6 2.5 3.9 1.1 versicolor71 5.9 3.2 4.8 1.8 versicolor72 6.1 2.8 4.0 1.3 versicolor73 6.3 2.5 4.9 1.5 versicolor74 6.1 2.8 4.7 1.2 versicolor75 6.4 2.9 4.3 1.3 versicolor76 6.6 3.0 4.4 1.4 versicolor77 6.8 2.8 4.8 1.4 versicolor78 6.7 3.0 5.0 1.7 versicolor79 6.0 2.9 4.5 1.5 versicolor80 5.7 2.6 3.5 1.0 versicolor81 5.5 2.4 3.8 1.1 versicolor82 5.5 2.4 3.7 1.0 versicolor83 5.8 2.7 3.9 1.2 versicolor84 6.0 2.7 5.1 1.6 versicolor85 5.4 3.0 4.5 1.5 versicolor86 6.0 3.4 4.5 1.6 versicolor87 6.7 3.1 4.7 1.5 versicolor88 6.3 2.3 4.4 1.3 versicolor89 5.6 3.0 4.1 1.3 versicolor90 5.5 2.5 4.0 1.3 versicolor91 5.5 2.6 4.4 1.2 versicolor92 6.1 3.0 4.6 1.4 versicolor93 5.8 2.6 4.0 1.2 versicolor94 5.0 2.3 3.3 1.0 versicolor95 5.6 2.7 4.2 1.3 versicolor96 5.7 3.0 4.2 1.2 versicolor97 5.7 2.9 4.2 1.3 versicolor98 6.2 2.9 4.3 1.3 versicolor99 5.1 2.5 3.0 1.1 versicolor100 5.7 2.8 4.1 1.3 versicolor101 6.3 3.3 6.0 2.5 virginica102 5.8 2.7 5.1 1.9 virginica103 7.1 3.0 5.9 2.1 virginica104 6.3 2.9 5.6 1.8 virginica105 6.5 3.0 5.8 2.2 virginica106 7.6 3.0 6.6 2.1 virginica107 4.9 2.5 4.5 1.7 virginica108 7.3 2.9 6.3 1.8 virginica109 6.7 2.5 5.8 1.8 virginica110 7.2 3.6 6.1 2.5 virginica111 6.5 3.2 5.1 2.0 virginica112 6.4 2.7 5.3 1.9 virginica113 6.8 3.0 5.5 2.1 virginica114 5.7 2.5 5.0 2.0 virginica115 5.8 2.8 5.1 2.4 virginica116 6.4 3.2 5.3 2.3 virginica117 6.5 3.0 5.5 1.8 virginica118 7.7 3.8 6.7 2.2 virginica119 7.7 2.6 6.9 2.3 virginica120 6.0 2.2 5.0 1.5 virginica121 6.9 3.2 5.7 2.3 virginica122 5.6 2.8 4.9 2.0 virginica123 7.7 2.8 6.7 2.0 virginica124 6.3 2.7 4.9 1.8 virginica125 6.7 3.3 5.7 2.1 virginica126 7.2 3.2 6.0 1.8 virginica127 6.2 2.8 4.8 1.8 virginica128 6.1 3.0 4.9 1.8 virginica129 6.4 2.8 5.6 2.1 virginica130 7.2 3.0 5.8 1.6 virginica131 7.4 2.8 6.1 1.9 virginica132 7.9 3.8 6.4 2.0 virginica133 6.4 2.8 5.6 2.2 virginica134 6.3 2.8 5.1 1.5 virginica135 6.1 2.6 5.6 1.4 virginica136 7.7 3.0 6.1 2.3 virginica137 6.3 3.4 5.6 2.4 virginica138 6.4 3.1 5.5 1.8 virginica139 6.0 3.0 4.8 1.8 virginica140 6.9 3.1 5.4 2.1 virginica141 6.7 3.1 5.6 2.4 virginica142 6.9 3.1 5.1 2.3 virginica143 5.8 2.7 5.1 1.9 virginica144 6.8 3.2 5.9 2.3 virginica145 6.7 3.3 5.7 2.5 virginica146 6.7 3.0 5.2 2.3 virginica147 6.3 2.5 5.0 1.9 virginica148 6.5 3.0 5.2 2.0 virginica149 6.2 3.4 5.4 2.3 virginica150 5.9 3.0 5.1 1.8 virginicaStructure of a list
str(first_list)List of 3 $ : int [1:3] 1 2 3 $ : int [1:2, 1:3] 1 2 3 4 5 6 $ :'data.frame': 150 obs. of 5 variables: ..$ Sepal.Length: num [1:150] 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ... ..$ Sepal.Width : num [1:150] 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 ... ..$ Petal.Length: num [1:150] 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 ... ..$ Petal.Width : num [1:150] 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ... ..$ Species : Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 ...Structure of a list
str(first_list)List of 3 $ : int [1:3] 1 2 3 $ : int [1:2, 1:3] 1 2 3 4 5 6 $ :'data.frame': 150 obs. of 5 variables: ..$ Sepal.Length: num [1:150] 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ... ..$ Sepal.Width : num [1:150] 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 ... ..$ Petal.Length: num [1:150] 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 ... ..$ Petal.Width : num [1:150] 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ... ..$ Species : Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 ...Extract elements
first_list[[1]][1] 1 2 3first_list[[3]]$Species [1] setosa setosa setosa setosa setosa setosa [7] setosa setosa setosa setosa setosa setosa [13] setosa setosa setosa setosa setosa setosa [19] setosa setosa setosa setosa setosa setosa [25] setosa setosa setosa setosa setosa setosa [31] setosa setosa setosa setosa setosa setosa [37] setosa setosa setosa setosa setosa setosa [43] setosa setosa setosa setosa setosa setosa [49] setosa setosa versicolor versicolor versicolor versicolor [55] versicolor versicolor versicolor versicolor versicolor versicolor [61] versicolor versicolor versicolor versicolor versicolor versicolor [67] versicolor versicolor versicolor versicolor versicolor versicolor [73] versicolor versicolor versicolor versicolor versicolor versicolor [79] versicolor versicolor versicolor versicolor versicolor versicolor [85] versicolor versicolor versicolor versicolor versicolor versicolor [91] versicolor versicolor versicolor versicolor versicolor versicolor [97] versicolor versicolor versicolor versicolor virginica virginica [103] virginica virginica virginica virginica virginica virginica [109] virginica virginica virginica virginica virginica virginica [115] virginica virginica virginica virginica virginica virginica [121] virginica virginica virginica virginica virginica virginica [127] virginica virginica virginica virginica virginica virginica [133] virginica virginica virginica virginica virginica virginica [139] virginica virginica virginica virginica virginica virginica [145] virginica virginica virginica virginica virginica virginica Levels: setosa versicolor virginicaName entries in a list
first_list_with_names <-list(a=1:3, b=matrix(1:6, nrow=2), c=iris)first_list_with_names$a[1] 1 2 3$b [,1] [,2] [,3][1,] 1 3 5[2,] 2 4 6$c Sepal.Length Sepal.Width Petal.Length Petal.Width Species1 5.1 3.5 1.4 0.2 setosa2 4.9 3.0 1.4 0.2 setosa3 4.7 3.2 1.3 0.2 setosa4 4.6 3.1 1.5 0.2 setosa5 5.0 3.6 1.4 0.2 setosa6 5.4 3.9 1.7 0.4 setosa7 4.6 3.4 1.4 0.3 setosa8 5.0 3.4 1.5 0.2 setosa9 4.4 2.9 1.4 0.2 setosa10 4.9 3.1 1.5 0.1 setosa11 5.4 3.7 1.5 0.2 setosa12 4.8 3.4 1.6 0.2 setosa13 4.8 3.0 1.4 0.1 setosa14 4.3 3.0 1.1 0.1 setosa15 5.8 4.0 1.2 0.2 setosa16 5.7 4.4 1.5 0.4 setosa17 5.4 3.9 1.3 0.4 setosa18 5.1 3.5 1.4 0.3 setosa19 5.7 3.8 1.7 0.3 setosa20 5.1 3.8 1.5 0.3 setosa21 5.4 3.4 1.7 0.2 setosa22 5.1 3.7 1.5 0.4 setosa23 4.6 3.6 1.0 0.2 setosa24 5.1 3.3 1.7 0.5 setosa25 4.8 3.4 1.9 0.2 setosa26 5.0 3.0 1.6 0.2 setosa27 5.0 3.4 1.6 0.4 setosa28 5.2 3.5 1.5 0.2 setosa29 5.2 3.4 1.4 0.2 setosa30 4.7 3.2 1.6 0.2 setosa31 4.8 3.1 1.6 0.2 setosa32 5.4 3.4 1.5 0.4 setosa33 5.2 4.1 1.5 0.1 setosa34 5.5 4.2 1.4 0.2 setosa35 4.9 3.1 1.5 0.2 setosa36 5.0 3.2 1.2 0.2 setosa37 5.5 3.5 1.3 0.2 setosa38 4.9 3.6 1.4 0.1 setosa39 4.4 3.0 1.3 0.2 setosa40 5.1 3.4 1.5 0.2 setosa41 5.0 3.5 1.3 0.3 setosa42 4.5 2.3 1.3 0.3 setosa43 4.4 3.2 1.3 0.2 setosa44 5.0 3.5 1.6 0.6 setosa45 5.1 3.8 1.9 0.4 setosa46 4.8 3.0 1.4 0.3 setosa47 5.1 3.8 1.6 0.2 setosa48 4.6 3.2 1.4 0.2 setosa49 5.3 3.7 1.5 0.2 setosa50 5.0 3.3 1.4 0.2 setosa51 7.0 3.2 4.7 1.4 versicolor52 6.4 3.2 4.5 1.5 versicolor53 6.9 3.1 4.9 1.5 versicolor54 5.5 2.3 4.0 1.3 versicolor55 6.5 2.8 4.6 1.5 versicolor56 5.7 2.8 4.5 1.3 versicolor57 6.3 3.3 4.7 1.6 versicolor58 4.9 2.4 3.3 1.0 versicolor59 6.6 2.9 4.6 1.3 versicolor60 5.2 2.7 3.9 1.4 versicolor61 5.0 2.0 3.5 1.0 versicolor62 5.9 3.0 4.2 1.5 versicolor63 6.0 2.2 4.0 1.0 versicolor64 6.1 2.9 4.7 1.4 versicolor65 5.6 2.9 3.6 1.3 versicolor66 6.7 3.1 4.4 1.4 versicolor67 5.6 3.0 4.5 1.5 versicolor68 5.8 2.7 4.1 1.0 versicolor69 6.2 2.2 4.5 1.5 versicolor70 5.6 2.5 3.9 1.1 versicolor71 5.9 3.2 4.8 1.8 versicolor72 6.1 2.8 4.0 1.3 versicolor73 6.3 2.5 4.9 1.5 versicolor74 6.1 2.8 4.7 1.2 versicolor75 6.4 2.9 4.3 1.3 versicolor76 6.6 3.0 4.4 1.4 versicolor77 6.8 2.8 4.8 1.4 versicolor78 6.7 3.0 5.0 1.7 versicolor79 6.0 2.9 4.5 1.5 versicolor80 5.7 2.6 3.5 1.0 versicolor81 5.5 2.4 3.8 1.1 versicolor82 5.5 2.4 3.7 1.0 versicolor83 5.8 2.7 3.9 1.2 versicolor84 6.0 2.7 5.1 1.6 versicolor85 5.4 3.0 4.5 1.5 versicolor86 6.0 3.4 4.5 1.6 versicolor87 6.7 3.1 4.7 1.5 versicolor88 6.3 2.3 4.4 1.3 versicolor89 5.6 3.0 4.1 1.3 versicolor90 5.5 2.5 4.0 1.3 versicolor91 5.5 2.6 4.4 1.2 versicolor92 6.1 3.0 4.6 1.4 versicolor93 5.8 2.6 4.0 1.2 versicolor94 5.0 2.3 3.3 1.0 versicolor95 5.6 2.7 4.2 1.3 versicolor96 5.7 3.0 4.2 1.2 versicolor97 5.7 2.9 4.2 1.3 versicolor98 6.2 2.9 4.3 1.3 versicolor99 5.1 2.5 3.0 1.1 versicolor100 5.7 2.8 4.1 1.3 versicolor101 6.3 3.3 6.0 2.5 virginica102 5.8 2.7 5.1 1.9 virginica103 7.1 3.0 5.9 2.1 virginica104 6.3 2.9 5.6 1.8 virginica105 6.5 3.0 5.8 2.2 virginica106 7.6 3.0 6.6 2.1 virginica107 4.9 2.5 4.5 1.7 virginica108 7.3 2.9 6.3 1.8 virginica109 6.7 2.5 5.8 1.8 virginica110 7.2 3.6 6.1 2.5 virginica111 6.5 3.2 5.1 2.0 virginica112 6.4 2.7 5.3 1.9 virginica113 6.8 3.0 5.5 2.1 virginica114 5.7 2.5 5.0 2.0 virginica115 5.8 2.8 5.1 2.4 virginica116 6.4 3.2 5.3 2.3 virginica117 6.5 3.0 5.5 1.8 virginica118 7.7 3.8 6.7 2.2 virginica119 7.7 2.6 6.9 2.3 virginica120 6.0 2.2 5.0 1.5 virginica121 6.9 3.2 5.7 2.3 virginica122 5.6 2.8 4.9 2.0 virginica123 7.7 2.8 6.7 2.0 virginica124 6.3 2.7 4.9 1.8 virginica125 6.7 3.3 5.7 2.1 virginica126 7.2 3.2 6.0 1.8 virginica127 6.2 2.8 4.8 1.8 virginica128 6.1 3.0 4.9 1.8 virginica129 6.4 2.8 5.6 2.1 virginica130 7.2 3.0 5.8 1.6 virginica131 7.4 2.8 6.1 1.9 virginica132 7.9 3.8 6.4 2.0 virginica133 6.4 2.8 5.6 2.2 virginica134 6.3 2.8 5.1 1.5 virginica135 6.1 2.6 5.6 1.4 virginica136 7.7 3.0 6.1 2.3 virginica137 6.3 3.4 5.6 2.4 virginica138 6.4 3.1 5.5 1.8 virginica139 6.0 3.0 4.8 1.8 virginica140 6.9 3.1 5.4 2.1 virginica141 6.7 3.1 5.6 2.4 virginica142 6.9 3.1 5.1 2.3 virginica143 5.8 2.7 5.1 1.9 virginica144 6.8 3.2 5.9 2.3 virginica145 6.7 3.3 5.7 2.5 virginica146 6.7 3.0 5.2 2.3 virginica147 6.3 2.5 5.0 1.9 virginica148 6.5 3.0 5.2 2.0 virginica149 6.2 3.4 5.4 2.3 virginica150 5.9 3.0 5.1 1.8 virginicaExtract elements using names
str(first_list_with_names)List of 3 $ a: int [1:3] 1 2 3 $ b: int [1:2, 1:3] 1 2 3 4 5 6 $ c:'data.frame': 150 obs. of 5 variables: ..$ Sepal.Length: num [1:150] 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ... ..$ Sepal.Width : num [1:150] 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 ... ..$ Petal.Length: num [1:150] 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 ... ..$ Petal.Width : num [1:150] 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ... ..$ Species : Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 ...first_list_with_names$a[1] 1 2 3first_list_with_names$c$Species [1] setosa setosa setosa setosa setosa setosa [7] setosa setosa setosa setosa setosa setosa [13] setosa setosa setosa setosa setosa setosa [19] setosa setosa setosa setosa setosa setosa [25] setosa setosa setosa setosa setosa setosa [31] setosa setosa setosa setosa setosa setosa [37] setosa setosa setosa setosa setosa setosa [43] setosa setosa setosa setosa setosa setosa [49] setosa setosa versicolor versicolor versicolor versicolor [55] versicolor versicolor versicolor versicolor versicolor versicolor [61] versicolor versicolor versicolor versicolor versicolor versicolor [67] versicolor versicolor versicolor versicolor versicolor versicolor [73] versicolor versicolor versicolor versicolor versicolor versicolor [79] versicolor versicolor versicolor versicolor versicolor versicolor [85] versicolor versicolor versicolor versicolor versicolor versicolor [91] versicolor versicolor versicolor versicolor versicolor versicolor [97] versicolor versicolor versicolor versicolor virginica virginica [103] virginica virginica virginica virginica virginica virginica [109] virginica virginica virginica virginica virginica virginica [115] virginica virginica virginica virginica virginica virginica [121] virginica virginica virginica virginica virginica virginica [127] virginica virginica virginica virginica virginica virginica [133] virginica virginica virginica virginica virginica virginica [139] virginica virginica virginica virginica virginica virginica [145] virginica virginica virginica virginica virginica virginica Levels: setosa versicolor virginicaYour turn
c("Jan","Feb","Mar"); matrix(c(3,9,5,1,-2,8), nrow = 2); list("green",12.3)[1] "Jan" "Feb" "Mar" [,1] [,2] [,3][1,] 3 5 -2[2,] 9 1 8[[1]][1] "green"[[2]][1] 12.3Create a list containing the above vector, matrix and the list.
Name the elements as
first,secondandthird.