STA 517 3.0 Programming and Statistical Computing with R
✍️ Regression Analysis
Dr Thiyanga Talagala
Today's menu
Fit a model
Visualise the fitted model
Measure the strength of the fit
Residual analysis
Make predictions

Packages
library(broom)library(modelr)library(GGally)library(carData)library(tidyverse)library(magrittr)library(car) # to calculate VIFData: Prestige of Canadian Occupations
head(Prestige, 5) education income women prestige census typegov.administrators 13.11 12351 11.16 68.8 1113 profgeneral.managers 12.26 25879 4.02 69.1 1130 profaccountants 12.77 9271 15.70 63.4 1171 profpurchasing.officers 11.42 8865 9.11 56.8 1175 profchemists 14.62 8403 11.68 73.5 2111 profsummary(Prestige) education income women prestige Min. : 6.380 Min. : 611 Min. : 0.000 Min. :14.80 1st Qu.: 8.445 1st Qu.: 4106 1st Qu.: 3.592 1st Qu.:35.23 Median :10.540 Median : 5930 Median :13.600 Median :43.60 Mean :10.738 Mean : 6798 Mean :28.979 Mean :46.83 3rd Qu.:12.648 3rd Qu.: 8187 3rd Qu.:52.203 3rd Qu.:59.27 Max. :15.970 Max. :25879 Max. :97.510 Max. :87.20 census type Min. :1113 bc :44 1st Qu.:3120 prof:31 Median :5135 wc :23 Mean :5402 NA's: 4 3rd Qu.:8312 Max. :9517Data description
prestige: prestige of Canadian occupations, measured by the Pineo-Porter prestige score for occupation taken from a social survey conducted in the mid-1960s.
education: Average education of occupational incumbents, years, in 1971.
income: Average income of incumbents, dollars, in 1971.
women: Percentage of incumbents who are women.
Data description (cont.)
census: Canadian Census occupational code.
type: Type of occupation.
- prof: professional and technical- wc: white collar- bc: blue collar- NA: missingThe dataset consists of 102 observations, each corresponding to a particular occupation.
Training test and Test set
## Create an ID per rowdf <- Prestige %>% mutate(id = row_number())## set the seed to make your partition reproducibleset.seed(123)train <- df %>% sample_frac(.80)dim(train)[1] 82 7test <- anti_join(df, train, by = 'id')dim(test)[1] 20 7Exploratory Data Analysis
train education income women prestige census type idmedical.technicians 12.79 5180 76.04 67.5 3156 wc 31welders 7.92 6477 5.17 41.8 8335 bc 79commercial.travellers 11.13 8780 3.16 40.2 5133 wc 51economists 14.44 8049 57.31 62.2 2311 prof 14farmers 6.84 3643 3.60 44.1 7112 <NA> 67receptionsts 11.04 2901 92.86 38.7 4171 wc 42sales.supervisors 9.84 7482 17.04 41.5 5130 wc 50mail.carriers 9.22 5511 7.62 36.1 4172 wc 43taxi.drivers 7.93 4224 3.59 25.1 9173 bc 99veterinarians 15.94 14558 4.32 66.7 3115 prof 25construction.foremen 8.24 8880 0.65 51.1 8780 bc 90carpenters 6.92 5299 0.56 38.9 8781 bc 91rotary.well.drillers 8.88 6860 0.00 35.3 7711 bc 69buyers 11.03 7956 23.88 51.1 5191 wc 57civil.engineers 14.52 11377 1.03 73.1 2143 prof 9slaughterers.2 7.64 5134 17.26 34.8 8215 bc 72osteopaths.chiropractors 14.71 17498 6.91 68.4 3117 prof 26biologists 15.09 8258 25.65 72.6 2133 prof 7train.engineers 8.49 8845 0.00 48.9 9131 bc 97electrical.linemen 9.05 8316 1.34 40.9 8731 bc 88typists 11.49 3148 95.97 41.9 4113 wc 36sheet.metal.workers 8.40 6565 2.30 35.9 8333 bc 78construction.labourers 7.52 3910 1.09 26.5 8798 bc 95tool.die.makers 10.09 8043 1.50 42.5 8311 bc 76psychologists 14.36 7405 48.28 74.9 2315 prof 15commercial.artists 11.09 6197 21.03 57.2 3314 prof 32auto.repairmen 8.10 5795 0.81 38.1 8581 bc 85radio.tv.repairmen 10.29 5449 2.92 37.2 8537 bc 83file.clerks 12.09 3016 83.19 32.7 4161 wc 41secondary.school.teachers 15.08 8034 46.80 66.1 2733 prof 23nurses 12.46 4614 96.12 64.7 3131 prof 27cooks 7.74 3116 52.00 29.7 6121 bc 60newsboys 9.62 918 7.00 14.8 5143 <NA> 53typesetters 10.00 6462 13.58 42.2 9511 bc 101bakers 7.54 4199 33.30 38.9 8213 bc 70railway.sectionmen 6.67 4696 0.00 27.3 8715 bc 87tellers.cashiers 10.64 2448 91.76 42.3 4133 wc 38athletes 11.44 8206 8.13 54.1 3373 <NA> 34physio.therapsts 13.62 5092 82.66 72.1 3137 prof 29chemists 14.62 8403 11.68 73.5 2111 prof 5architects 15.44 14163 2.69 78.1 2141 prof 8draughtsmen 12.30 7059 7.83 60.0 2163 prof 12computer.programers 13.83 8425 15.33 53.8 2183 prof 13librarians 14.15 6112 77.10 58.1 2351 prof 18radio.tv.announcers 12.71 7562 11.15 57.6 3337 wc 33electricians 9.93 7147 0.99 50.2 8733 bc 89bus.drivers 7.58 5562 9.47 35.9 9171 bc 98house.painters 7.81 4549 2.46 29.9 8785 bc 93elevator.operators 7.58 3582 30.08 20.1 6193 bc 66university.teachers 15.97 12480 19.59 84.6 2711 prof 21aircraft.workers 8.78 6573 5.78 43.7 8515 bc 81textile.weavers 6.69 4443 31.36 33.3 8267 bc 74claim.adjustors 11.13 5052 56.10 51.1 4192 wc 47aircraft.repairmen 10.10 7716 0.78 50.3 8582 bc 86bookbinders 8.55 3617 70.87 35.2 9517 bc 102social.workers 14.21 6336 54.77 55.1 2331 prof 16pharmacists 15.21 10432 24.71 69.3 3151 prof 30physicists 15.64 11030 5.13 77.6 2113 prof 6auto.workers 8.43 5811 13.62 35.9 8513 bc 80funeral.directors 10.57 7869 6.01 54.9 6141 bc 62primary.school.teachers 13.62 5648 83.78 59.6 2731 prof 22sewing.mach.operators 6.38 2847 90.67 28.2 8563 bc 84computer.operators 11.36 4330 75.92 47.7 4143 wc 39travel.clerks 11.43 6259 39.17 35.7 4193 wc 48lawyers 15.77 19263 5.13 82.3 2343 prof 17janitors 7.11 3472 33.57 17.3 6191 bc 65purchasing.officers 11.42 8865 9.11 56.8 1175 prof 4slaughterers.1 7.64 5134 17.26 25.2 8215 bc 71babysitters 9.46 611 96.53 25.9 6147 <NA> 63insurance.agents 11.60 8131 13.09 47.3 5171 wc 55ministers 14.50 4686 4.14 72.8 2511 prof 20longshoremen 8.37 4753 0.00 26.1 9313 bc 100firefighters 9.47 8895 0.00 43.5 6111 bc 58plumbers 8.33 6928 0.61 42.9 8791 bc 94collectors 11.20 4741 47.06 29.4 4191 wc 46canners 7.42 1890 72.24 23.2 8221 bc 73accountants 12.77 9271 15.70 63.4 1171 prof 3postal.clerks 10.07 3739 52.27 37.2 4173 wc 44pilots 12.27 14032 0.58 66.1 9111 prof 96bartenders 8.50 3930 15.51 20.2 6123 bc 61launderers 7.33 3000 69.31 20.8 6162 bc 64office.clerks 11.00 4075 63.23 35.6 4197 wc 49]
Prestige_1 <- train %>% pivot_longer(c(1, 2, 3, 4), names_to="variable", values_to="value")Prestige_1# A tibble: 328 × 5 census type id variable value <int> <fct> <int> <chr> <dbl> 1 3156 wc 31 education 12.8 2 3156 wc 31 income 5180 3 3156 wc 31 women 76.0 4 3156 wc 31 prestige 67.5 5 8335 bc 79 education 7.92 6 8335 bc 79 income 6477 7 8335 bc 79 women 5.17 8 8335 bc 79 prestige 41.8 9 5133 wc 51 education 11.1 10 5133 wc 51 income 8780 # … with 318 more rowsggplot(Prestige_1, aes(x=value)) + geom_histogram() + facet_wrap(variable ~., ncol=1)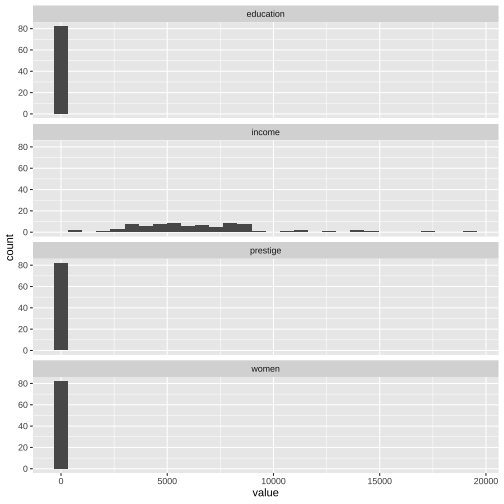
ggplot(Prestige_1, aes(x=value)) + geom_histogram() + facet_wrap(variable ~., ncol=1, scales = "free")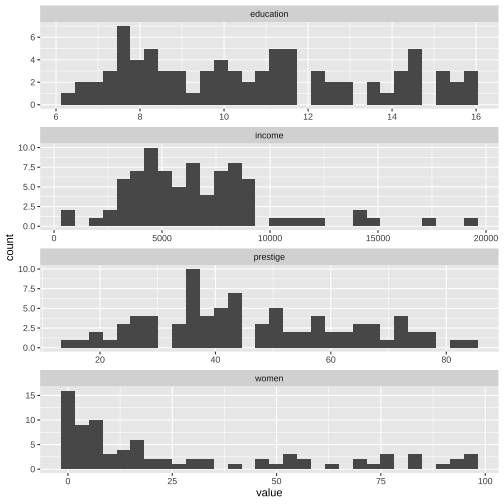
ggplot(Prestige_1, aes(x=value)) + geom_histogram(colour="white") + facet_wrap(variable ~., ncol=1, scales = "free")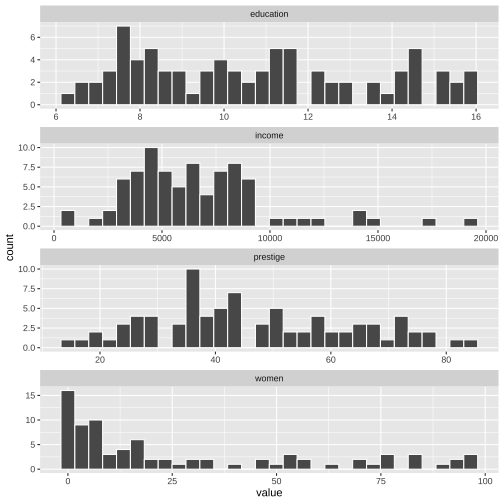
ggplot(Prestige_1, aes(x=value, fill=variable)) + geom_density() + facet_wrap(variable ~., ncol=1, scales = "free")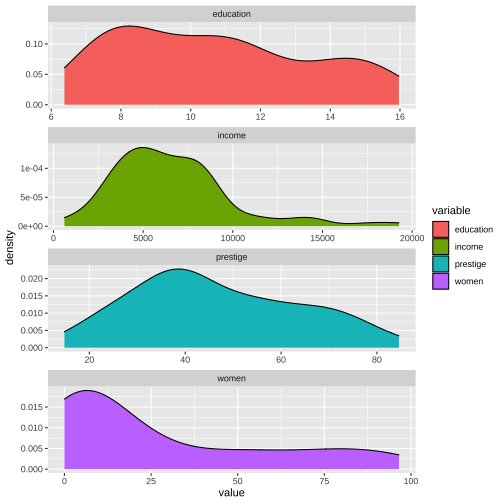
ggplot(Prestige_1, aes(y = value, x = type, fill = type)) + geom_boxplot() + facet_wrap(variable ~., ncol=1, scales = "free")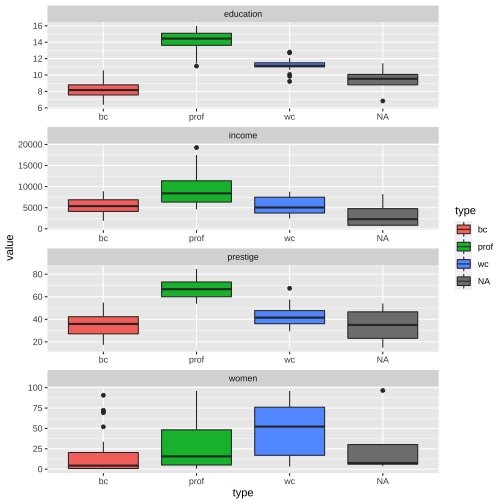
Prestige_1 %>% filter(is.na(type) == FALSE) %>%ggplot(aes(y=value, x=type, fill=type)) + geom_boxplot() + facet_wrap(variable ~., ncol=1, scales = "free")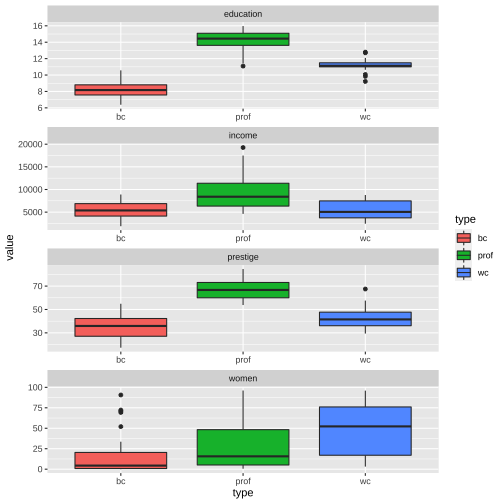
Prestige_1 %>% filter(is.na(type) == FALSE) %>%ggplot(aes(x = value, y = type, fill = type)) + geom_boxplot() + facet_wrap(variable ~., ncol=1, scales = "free")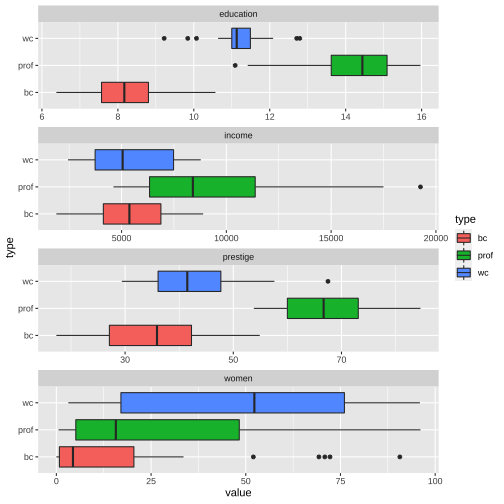
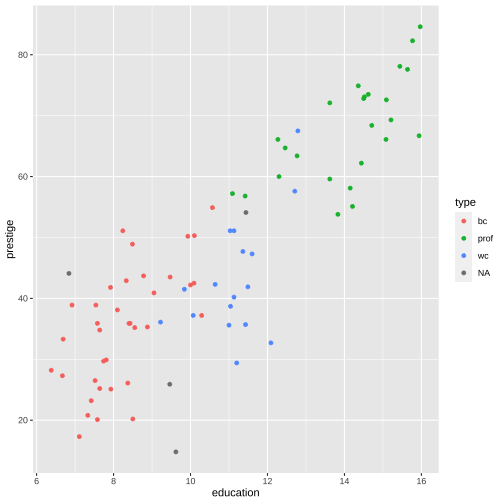
train %>% select(education, income, prestige, women) %>% ggpairs()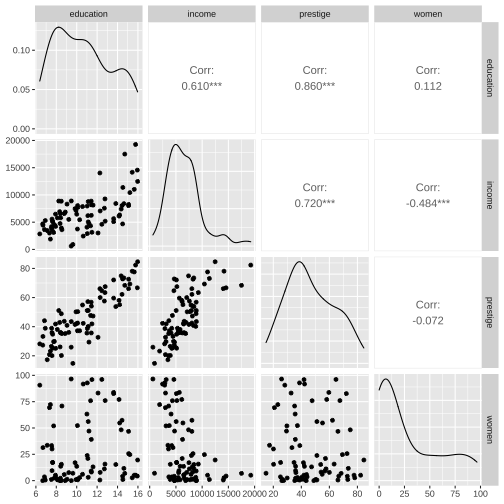
train %>% filter(is.na(type) == FALSE) %>% ggpairs(columns= c("education", "income", "prestige", "women"), mapping=aes(color=type))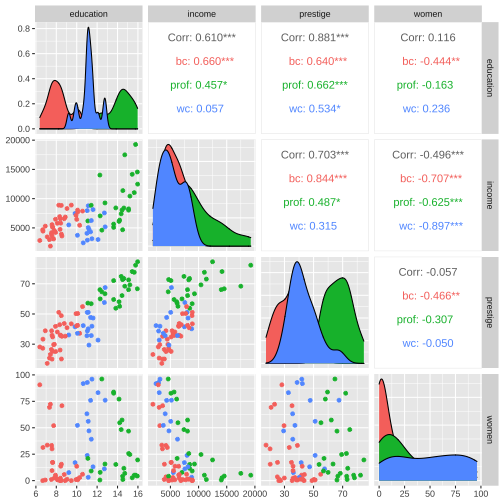
Regression analysis
Steps
Fit a model.
Visualize the fitted model.
Measuring the strength of the fit.
Residual analysis.
Interpret the coefficients.
Make predictions using the fitted model.
Model 1: prestige ~ education
Recap
True relationship between X and Y in the population
Y=f(X)+ϵ
If f is approximated by a linear function
Y=β0+β1X+ϵ
The error terms are normally distributed with mean 0 and variance σ2. Then the mean response, Y, at any value of the X is
E(Y|X=xi)=E(β0+β1xi+ϵ)=β0+β1xi
For a single unit (yi,xi)
yi=β0+β1xi+ϵi where ϵi∼N(0,σ2)
We use sample values (yi,xi) where i=1,2,...n to estimate β0 and β1.
The fitted regression model is
^Yi=^β0+^β1xi
How to estimate β0 and β1?
Sum of squares of Residuals
SSR=e21+e22+...+e2n
The least-squares regression approach chooses coefficients ^β0 and ^β1 to minimize SSR.
Model 1: prestige ~ education
1. Fit a model
Model 1: Fit a model
yi=β0+β1xi+ϵi,where ϵi∼NID(0,σ2)
To estimate β0 and β1
model1 <- lm(prestige ~ education, data=train)summary(model1)Call:lm(formula = prestige ~ education, data = train)Residuals: Min 1Q Median 3Q Max -26.1683 -6.1403 0.8302 6.2665 17.8892 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) -10.0990 3.8922 -2.595 0.0113 * education 5.3085 0.3519 15.085 <2e-16 ***---Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Residual standard error: 8.81 on 80 degrees of freedomMultiple R-squared: 0.7399, Adjusted R-squared: 0.7366 F-statistic: 227.5 on 1 and 80 DF, p-value: < 2.2e-16What's messy about the output?
Call:lm(formula = prestige ~ education, data = train)Residuals: Min 1Q Median 3Q Max -26.1683 -6.1403 0.8302 6.2665 17.8892 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) -10.0990 3.8922 -2.595 0.0113 * education 5.3085 0.3519 15.085 <2e-16 ***---Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Residual standard error: 8.81 on 80 degrees of freedomMultiple R-squared: 0.7399, Adjusted R-squared: 0.7366 F-statistic: 227.5 on 1 and 80 DF, p-value: < 2.2e-16Extract coefficients takes multiple steps.
data.frame(coef(summary(model1)))Column names are inconvenient to use.
Information are stored in row names.
broom functions
broom takes model objects and turns them into tidy data frames that can be used with other tidy tools.
Three main functions
tidy(): component-level statisticsaugment(): observation-level statisticsglance(): model-level statistics
Component-level statistics: tidy()
model1 %>% tidy()# A tibble: 2 × 5 term estimate std.error statistic p.value <chr> <dbl> <dbl> <dbl> <dbl>1 (Intercept) -10.1 3.89 -2.59 1.13e- 22 education 5.31 0.352 15.1 4.17e-25model1 %>% tidy(conf.int=TRUE)# A tibble: 2 × 7 term estimate std.error statistic p.value conf.low conf.high <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>1 (Intercept) -10.1 3.89 -2.59 1.13e- 2 -17.8 -2.352 education 5.31 0.352 15.1 4.17e-25 4.61 6.01Component-level statistics: tidy()
model1 %>% tidy() %>% select(term, estimate)# A tibble: 2 × 2 term estimate <chr> <dbl>1 (Intercept) -10.1 2 education 5.31Component-level statistics: tidy() (cont.)
model1 %>% tidy()# A tibble: 2 × 5 term estimate std.error statistic p.value <chr> <dbl> <dbl> <dbl> <dbl>1 (Intercept) -10.1 3.89 -2.59 1.13e- 22 education 5.31 0.352 15.1 4.17e-25Fitted model is
^Yi=−9.42+5.27xi
Why are tidy model outputs useful?
tidy_model1 <- model1 %>% tidy(conf.int=TRUE)ggplot(tidy_model1, aes(x=estimate, y=term, color=term)) + geom_point() + geom_errorbarh(aes(xmin = conf.low, xmax=conf.high))+ggtitle("Coefficient plot")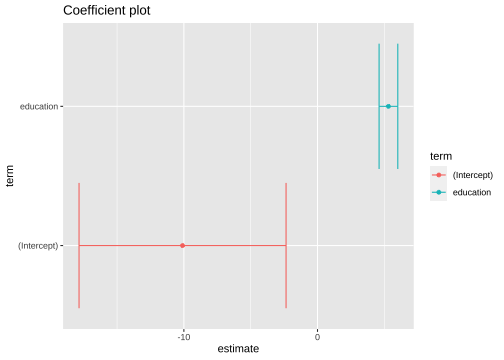
Model 1: prestige ~ education
1. Fit a model
2. Visualise the fitted model
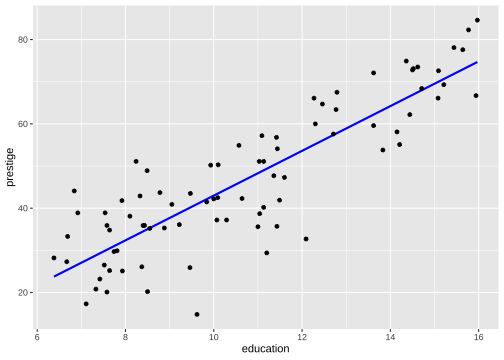
Model 1: Visualise the fitted model
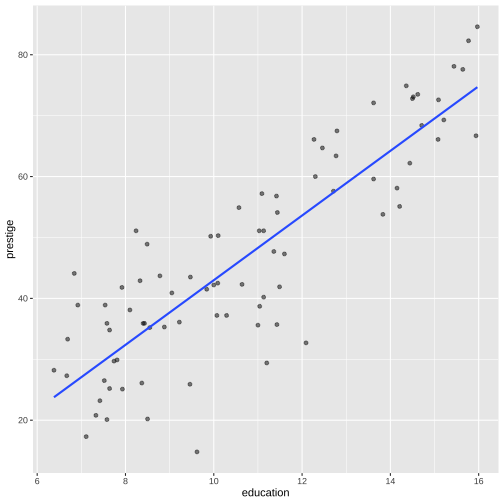
Model 1: Visualise the fitted model (style the line)
ggplot(data=train, aes(y=prestige, x=education)) + geom_point(alpha=0.5) + geom_smooth(method="lm", se=FALSE, col="forestgreen", lwd=2)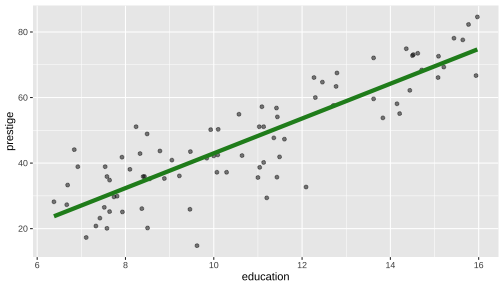
Model 1: prestige ~ education
1. Fit a model
2. Visualise the fitted model
3. Measure the strength of the fit
Model-level statistics: glance()
Measuring the strength of the fit
glance(model1)# A tibble: 1 × 12 r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>1 0.740 0.737 8.81 228. 4.17e-25 1 -294. 594. 601.# … with 3 more variables: deviance <dbl>, df.residual <int>, nobs <int>glance(model1)$adj.r.squared # extract values[1] 0.7366237Roughly 73% of the variability in prestige can be explained by the variable education.
Model 1: prestige ~ education
1. Fit a model
2. Visualise the fitted model
3. Measure the strength of the fit
4. Residual analysis
Observation-level statistics: augment()
model1_fitresid <- augment(model1)model1_fitresid# A tibble: 82 × 9 .rownames prestige education .fitted .resid .hat .sigma .cooksd .std.resid <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> 1 medical.t… 67.5 12.8 57.8 9.70 0.0191 8.80 1.20e-2 1.11 2 welders 41.8 7.92 31.9 9.86 0.0246 8.79 1.62e-2 1.13 3 commercia… 40.2 11.1 49.0 -8.78 0.0125 8.81 6.36e-3 -1.00 4 economists 62.2 14.4 66.6 -4.36 0.0344 8.85 4.51e-3 -0.503 5 farmers 44.1 6.84 26.2 17.9 0.0361 8.63 8.01e-2 2.07 6 reception… 38.7 11.0 48.5 -9.81 0.0124 8.80 7.86e-3 -1.12 7 sales.sup… 41.5 9.84 42.1 -0.636 0.0134 8.87 3.59e-5 -0.0727 8 mail.carr… 36.1 9.22 38.8 -2.74 0.0157 8.86 7.88e-4 -0.314 9 taxi.driv… 25.1 7.93 32.0 -6.90 0.0245 8.83 7.90e-3 -0.793 10 veterinar… 66.7 15.9 74.5 -7.82 0.0559 8.82 2.47e-2 -0.913 # … with 72 more rowsResiduals and Fitted Values
Residuals and Fitted Values
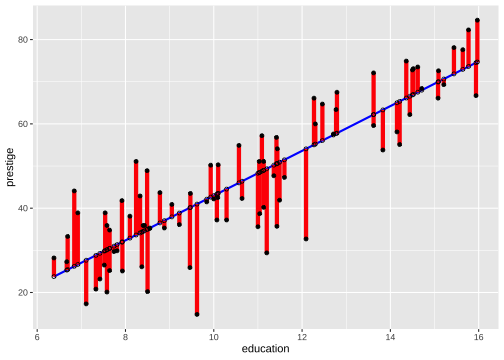
The residual is the difference between the observed and predicted response.
The residual for the ith observation is
ei=yi−^Yi=yi−(^β0+^β1xi)
Conditions for inference for regression
The relationship between the response and the predictors is linear.
The error terms are assumed to have zero mean and unknown constant variance σ2.
The errors are normally distributed.
The errors are uncorrelated.
Plot of residuals in time sequence.
The errors are uncorrelated.
Often, we can conclude that the this assumption is sufficiently met based on a description of the data and how it was collected.
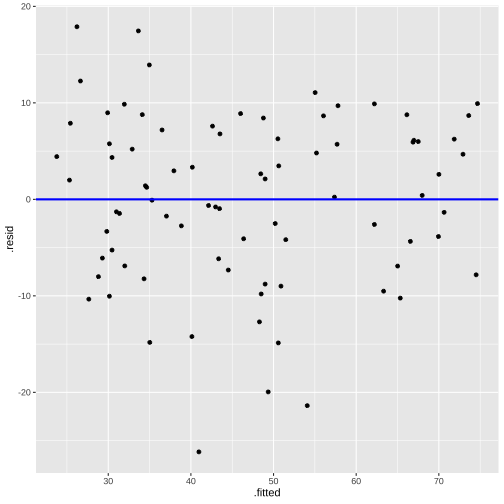
Plot of residuals vs fitted values
This is useful for detecting several common types of model inadequacies.
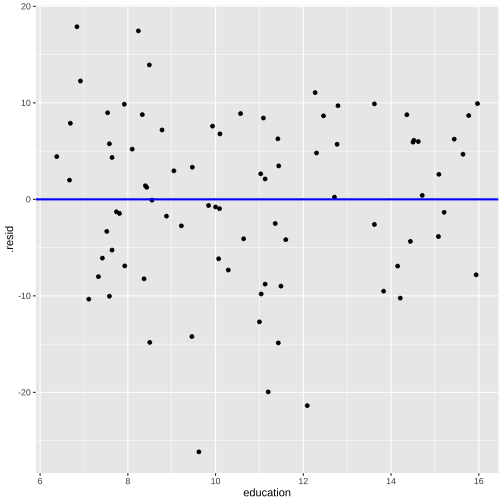
Plot of residuals vs fitted values and Plot of residuals vs predictor
linearity and constant variance
Residuals vs Fitted
ggplot(model1_fitresid, aes(x = .fitted, y = .resid))+ ------ + ------Residuals vs X
ggplot(model1_fitresid, aes(x = education, y = .resid))+ ------ + ------Residuals vs Fitted
Residuals vs X
Normality of residuals
ggplot(model1_fitresid, aes(x=.resid))+ geom_histogram(colour="white")+ggtitle("Distribution of Residuals")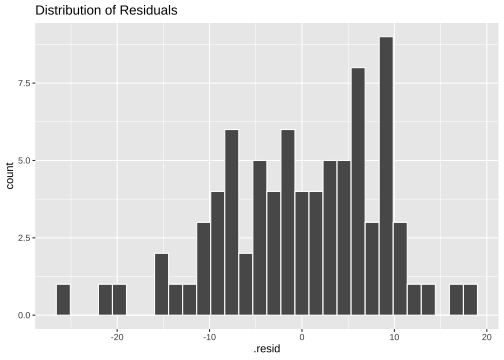
ggplot(model1_fitresid, aes(sample=.resid))+ stat_qq() + stat_qq_line()+labs(x="Theoretical Quantiles", y="Sample Quantiles")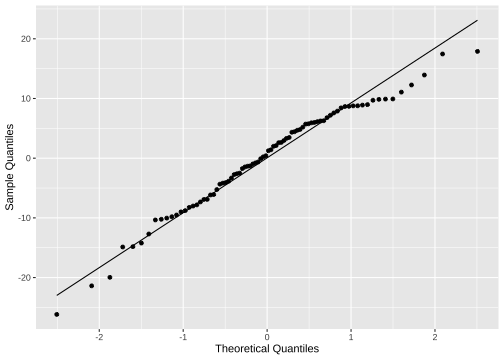
shapiro.test(model1_fitresid$.resid) Shapiro-Wilk normality testdata: model1_fitresid$.residW = 0.97547, p-value = 0.1176Model 2: prestige ~ education + income
1. Fit a model
2. Visualise the fitted model
3. Measure the strength of the fit
4. Residual analysis
Model 2: prestige ~ education + income
model2 <- lm(prestige ~ income + education, data=train)summary(model2)Call:lm(formula = prestige ~ income + education, data = train)Residuals: Min 1Q Median 3Q Max -18.4585 -5.1455 0.0464 5.0861 18.0320 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) -7.9953726 3.4549442 -2.314 0.0233 * income 0.0015822 0.0003222 4.911 4.79e-06 ***education 4.1373699 0.3910785 10.579 < 2e-16 ***---Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Residual standard error: 7.76 on 79 degrees of freedomMultiple R-squared: 0.8007, Adjusted R-squared: 0.7957 F-statistic: 158.7 on 2 and 79 DF, p-value: < 2.2e-16Model 2: prestige ~ education + income (cont.)
model2_fitresid <- augment(model2)model2_fitresid# A tibble: 82 × 10 .rownames prestige income education .fitted .resid .hat .sigma .cooksd <chr> <dbl> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> 1 medical.techn… 67.5 5180 12.8 53.1 14.4 0.0342 7.63 4.20e-2 2 welders 41.8 6477 7.92 35.0 6.78 0.0311 7.77 8.44e-3 3 commercial.tr… 40.2 8780 11.1 51.9 -11.7 0.0185 7.69 1.47e-2 4 economists 62.2 8049 14.4 64.5 -2.28 0.0374 7.80 1.16e-3 5 farmers 44.1 3643 6.84 26.1 18.0 0.0361 7.53 6.99e-2 6 receptionsts 38.7 2901 11.0 42.3 -3.57 0.0391 7.80 2.99e-3 7 sales.supervi… 41.5 7482 9.84 44.6 -3.05 0.0174 7.80 9.32e-4 8 mail.carriers 36.1 5511 9.22 38.9 -2.77 0.0157 7.80 6.90e-4 9 taxi.drivers 25.1 4224 7.93 31.5 -6.40 0.0247 7.77 5.88e-310 veterinarians 66.7 14558 15.9 81.0 -14.3 0.0847 7.62 1.14e-1# … with 72 more rows, and 1 more variable: .std.resid <dbl>Plot of residuals vs fitted values
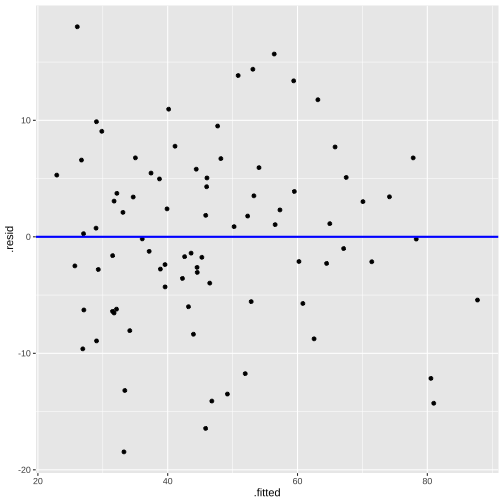
linearity and constant variance?
Normality of residuals
ggplot(model2_fitresid, aes(x=.resid))+ geom_histogram(colour="white")+ggtitle("Distribution of Residuals")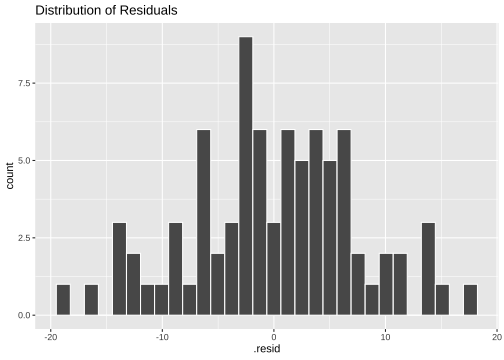
ggplot(model2_fitresid, aes(sample=.resid))+ stat_qq() + stat_qq_line()+labs(x="Theoretical Quantiles", y="Sample Quantiles")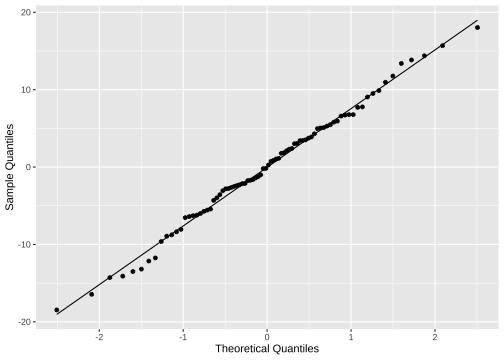
shapiro.test(model2_fitresid$.resid) Shapiro-Wilk normality testdata: model2_fitresid$.residW = 0.99298, p-value = 0.9405Plot of residuals vs predictor variables
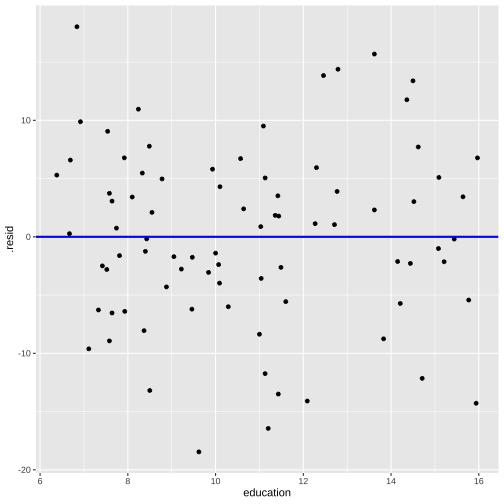
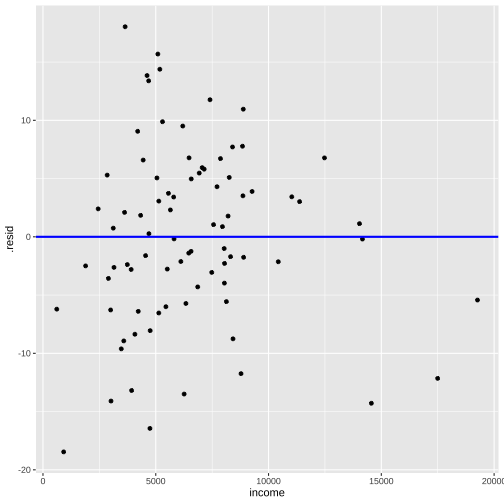
Model 3: prestige ~ education + log(income)
1. Fit a model
2. Visualise the fitted model
3. Measure the strength of the fit
4. Residual analysis
Model 3: prestige ~ education + log(income)
model3 <- lm(prestige ~ log(income) + education, data=train)summary(model3)Call:lm(formula = prestige ~ log(income) + education, data = train)Residuals: Min 1Q Median 3Q Max -17.3538 -4.7221 0.7046 4.3980 18.4684 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) -89.0742 13.2673 -6.714 2.62e-09 ***log(income) 10.4746 1.7069 6.136 3.16e-08 ***education 4.2117 0.3419 12.320 < 2e-16 ***---Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Residual standard error: 7.296 on 79 degrees of freedomMultiple R-squared: 0.8238, Adjusted R-squared: 0.8194 F-statistic: 184.7 on 2 and 79 DF, p-value: < 2.2e-16model3_fitresid <- augment(model3)model3_fitresid# A tibble: 82 × 9 .rownames prestige `log(income)` education .fitted .hat .sigma .cooksd <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> 1 medical.techn… 67.5 8.55 12.8 54.4 0.0249 7.19 0.0283 2 welders 41.8 8.78 7.92 36.2 0.0337 7.31 0.00706 3 commercial.tr… 40.2 9.08 11.1 52.9 0.0202 7.20 0.0213 4 economists 62.2 8.99 14.4 65.9 0.0346 7.33 0.00326 5 farmers 44.1 8.20 6.84 25.6 0.0362 7.03 0.0834 6 receptionsts 38.7 7.97 11.0 40.9 0.0410 7.34 0.00139 7 sales.supervi… 41.5 8.92 9.84 45.8 0.0201 7.33 0.00243 8 mail.carriers 36.1 8.61 9.22 40.0 0.0164 7.33 0.00161 9 taxi.drivers 25.1 8.35 7.93 31.8 0.0245 7.30 0.0071910 veterinarians 66.7 9.59 15.9 78.5 0.0636 7.21 0.0630 # … with 72 more rows, and 1 more variable: .std.resid <dbl>If the variables used to fit the model are not included in data, then no .resid column will be included in the output.
When you transform variables (say a log transformation), augment will not display .resid column.
Add new variable (log.income) to the training set.
train$log.income. <- log(train$income)model3 <- lm(prestige ~ log.income. + education, data=train)model3_fitresid <- broom::augment(model3)model3_fitresid# A tibble: 82 × 10 .rownames prestige log.income. education .fitted .resid .hat .sigma .cooksd <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> 1 medical.… 67.5 8.55 12.8 54.4 13.1 0.0249 7.19 0.0283 2 welders 41.8 8.78 7.92 36.2 5.59 0.0337 7.31 0.00706 3 commerci… 40.2 9.08 11.1 52.9 -12.7 0.0202 7.20 0.0213 4 economis… 62.2 8.99 14.4 65.9 -3.74 0.0346 7.33 0.00326 5 farmers 44.1 8.20 6.84 25.6 18.5 0.0362 7.03 0.0834 6 receptio… 38.7 7.97 11.0 40.9 -2.23 0.0410 7.34 0.00139 7 sales.su… 41.5 8.92 9.84 45.8 -4.31 0.0201 7.33 0.00243 8 mail.car… 36.1 8.61 9.22 40.0 -3.89 0.0164 7.33 0.00161 9 taxi.dri… 25.1 8.35 7.93 31.8 -6.67 0.0245 7.30 0.0071910 veterina… 66.7 9.59 15.9 78.5 -11.8 0.0636 7.21 0.0630 # … with 72 more rows, and 1 more variable: .std.resid <dbl>Plot of Residuals vs Fitted
Now - Model 3
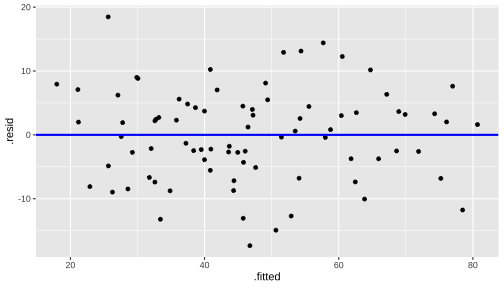
Before - Model 2
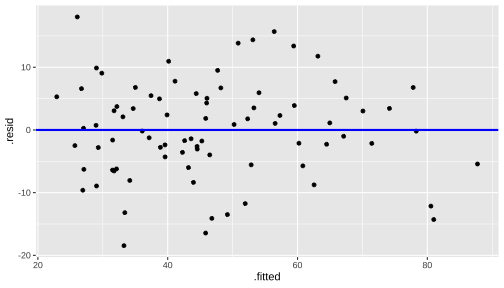
Normality of residuals
ggplot(model3_fitresid, aes(x=.resid))+ geom_histogram(colour="white")+ggtitle("Distribution of Residuals")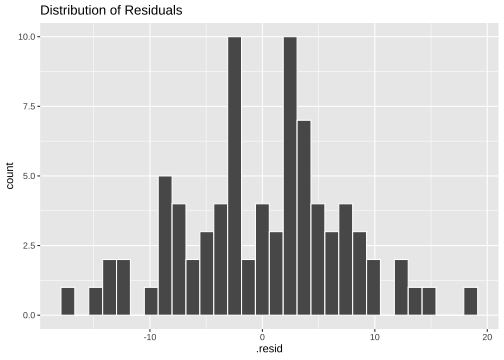
ggplot(model3_fitresid, aes(sample=.resid))+ stat_qq() + stat_qq_line()+labs(x="Theoretical Quantiles", y="Sample Quantiles")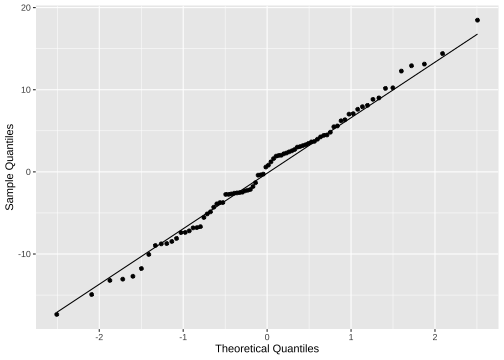
shapiro.test(model3_fitresid$.resid) Shapiro-Wilk normality testdata: model3_fitresid$.residW = 0.99414, p-value = 0.9747Plot of residuals vs predictor variables
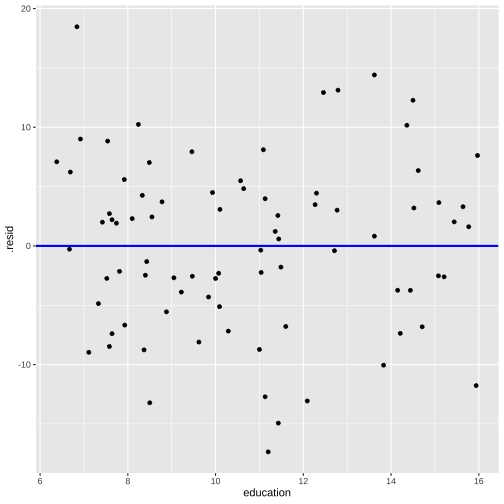
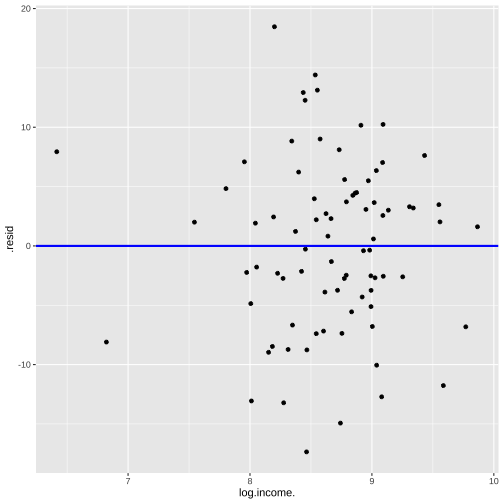
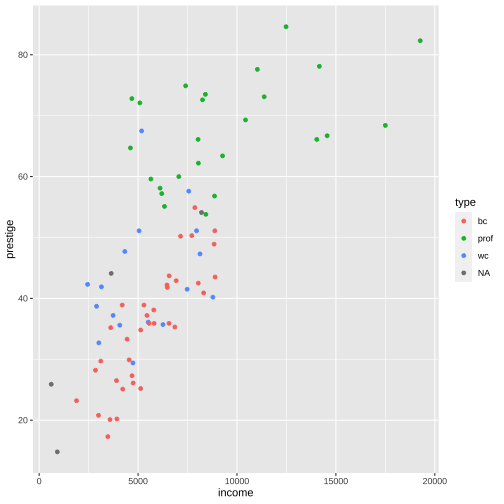
Prestige vs income by type
R code: _
Prestige vs income by type
R code: __
Model 4: prestige ~ education + log(income) + type
1. Fit a model
2. Visualise the fitted model
3. Measure the strength of the fit
4. Residual analysis
str(train)'data.frame': 82 obs. of 8 variables: $ education : num 12.79 7.92 11.13 14.44 6.84 ... $ income : int 5180 6477 8780 8049 3643 2901 7482 5511 4224 14558 ... $ women : num 76.04 5.17 3.16 57.31 3.6 ... $ prestige : num 67.5 41.8 40.2 62.2 44.1 38.7 41.5 36.1 25.1 66.7 ... $ census : int 3156 8335 5133 2311 7112 4171 5130 4172 9173 3115 ... $ type : Factor w/ 3 levels "bc","prof","wc": 3 1 3 2 NA 3 3 3 1 2 ... $ id : int 31 79 51 14 67 42 50 43 99 25 ... $ log.income.: num 8.55 8.78 9.08 8.99 8.2 ...Model 4: prestige ~ education + log(income) + type
model4 <- lm(prestige ~ log.income. + education + type, data=train)summary(model4)Call:lm(formula = prestige ~ log.income. + education + type, data = train)Residuals: Min 1Q Median 3Q Max -13.4996 -4.5672 0.5835 4.7882 18.1563 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) -75.5417 17.4258 -4.335 4.59e-05 ***log.income. 9.5470 2.2593 4.226 6.80e-05 ***education 3.5211 0.7523 4.681 1.29e-05 ***typeprof 6.6951 4.3625 1.535 0.129 typewc -1.8008 2.9820 -0.604 0.548 ---Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Residual standard error: 6.721 on 73 degrees of freedom (4 observations deleted due to missingness)Multiple R-squared: 0.8523, Adjusted R-squared: 0.8442 F-statistic: 105.3 on 4 and 73 DF, p-value: < 2.2e-16Model 4: prestige ~ education + log(income) + type
model4_fitresid <- augment(model4)head(model4_fitresid)# A tibble: 6 × 11 .rownames prestige log.income. education type .fitted .resid .hat .sigma <chr> <dbl> <dbl> <dbl> <fct> <dbl> <dbl> <dbl> <dbl>1 medical.tec… 67.5 8.55 12.8 wc 49.3 18.2 0.0895 6.382 welders 41.8 8.78 7.92 bc 36.1 5.67 0.0373 6.733 commercial.… 40.2 9.08 11.1 wc 48.5 -8.34 0.0970 6.694 economists 62.2 8.99 14.4 prof 67.9 -5.66 0.0431 6.735 receptionsts 38.7 7.97 11.0 wc 37.6 1.05 0.0887 6.776 sales.super… 41.5 8.92 9.84 wc 42.5 -0.967 0.119 6.77# … with 2 more variables: .cooksd <dbl>, .std.resid <dbl>Plot of Residuals vs Fitted
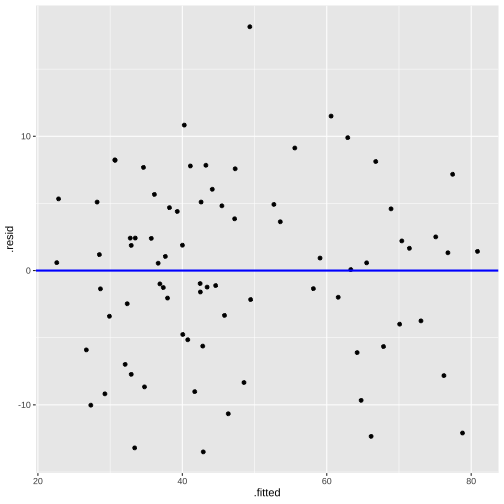
Normality of residuals
ggplot(model4_fitresid, aes(x=.resid))+ geom_histogram(colour="white")+ggtitle("Distribution of Residuals")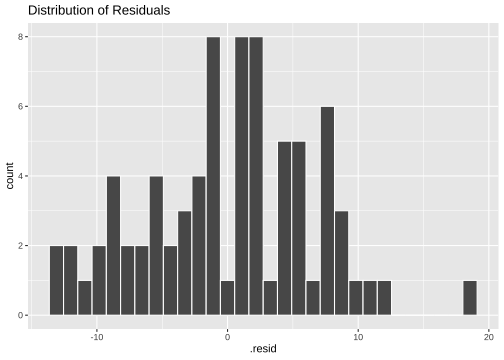
ggplot(model4_fitresid, aes(sample=.resid))+ stat_qq() + stat_qq_line()+labs(x="Theoretical Quantiles", y="Sample Quantiles")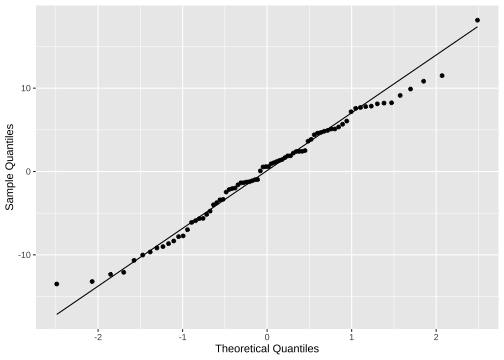
shapiro.test(model4_fitresid$.resid) Shapiro-Wilk normality testdata: model4_fitresid$.residW = 0.98621, p-value = 0.5658Plot of residuals vs predictor variables
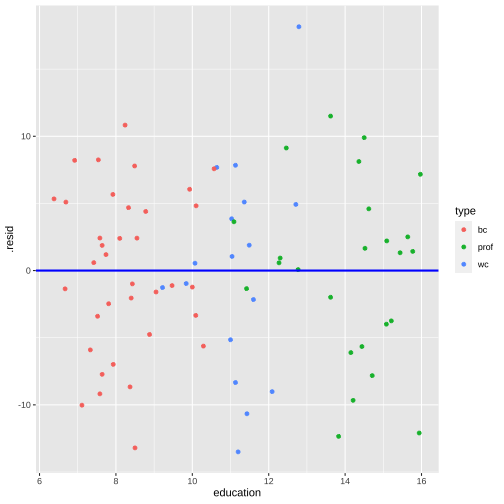
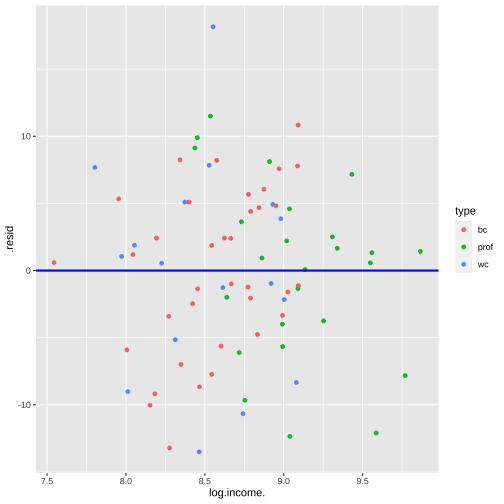
Multicollinearity
car::vif(model4) GVIF Df GVIF^(1/(2*Df))log.income. 1.825493 1 1.351108education 7.618522 1 2.760167type 7.159248 2 1.635750VIFs larger than 10 imply series problems with multicollinearity.
Detecting influential observations
library(lindia)gg_cooksd(model4)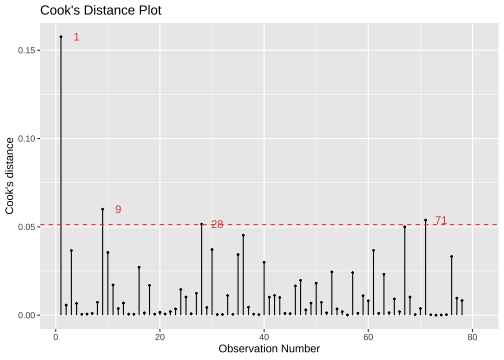
Influential outliers (cont.)
model4_fitresid %>% top_n(4, wt = .cooksd)# A tibble: 4 × 11 .rownames prestige log.income. education type .fitted .resid .hat .sigma <chr> <dbl> <dbl> <dbl> <fct> <dbl> <dbl> <dbl> <dbl>1 medical.tec… 67.5 8.55 12.8 wc 49.3 18.2 0.0895 6.382 veterinaria… 66.7 9.59 15.9 prof 78.8 -12.1 0.0786 6.603 file.clerks 32.7 8.01 12.1 wc 41.7 -9.02 0.113 6.674 collectors 29.4 8.46 11.2 wc 42.9 -13.5 0.0591 6.57# … with 2 more variables: .cooksd <dbl>, .std.resid <dbl>Solutions
Remove influential observations, and re-fit model.
Transform explanatory variables to reduce influence.
Use weighted regression to down weight influence of extreme observations.
Hypothesis testing
Y=β0+β1x1+β2x2+β3x3+β4x4+ϵ
What is the overall adequacy of the model?
H0:β1=β2=β3=β4=0
H1:βj≠0 for at least one j,j=1,2,3,4
Which specific regressors seem important?
H0:β1=0
H1:β1≠0
Making predictions
Method 1
head(test) education income women prestige census type idgov.administrators 13.11 12351 11.16 68.8 1113 prof 1general.managers 12.26 25879 4.02 69.1 1130 prof 2mining.engineers 14.64 11023 0.94 68.8 2153 prof 10surveyors 12.39 5902 1.91 62.0 2161 prof 11vocational.counsellors 15.22 9593 34.89 58.3 2391 prof 19physicians 15.96 25308 10.56 87.2 3111 prof 24test$log.income. <- log(test$income)predict(model4, test) gov.administrators general.managers mining.engineers 67.26199 71.33086 71.56331 surveyors vocational.counsellors physicians 57.67687 72.27901 84.14602 nursing.aides secretaries bookkeepers 35.60007 42.73588 42.49606 shipping.clerks telephone.operators sales.clerks 35.79196 36.60012 33.09309 service.station.attendant real.estate.salesmen policemen 33.60911 46.22149 49.75274 farm.workers textile.labourers machinists 25.50358 26.05783 39.56685 electronic.workers masons 34.34687 30.68619Making predictions
Method 2
library(modelr)test <- test %>% add_predictions(model4)head(test, 4) education income women prestige census type id log.income.gov.administrators 13.11 12351 11.16 68.8 1113 prof 1 9.421492general.managers 12.26 25879 4.02 69.1 1130 prof 2 10.161187mining.engineers 14.64 11023 0.94 68.8 2153 prof 10 9.307739surveyors 12.39 5902 1.91 62.0 2161 prof 11 8.683047 predgov.administrators 67.26199general.managers 71.33086mining.engineers 71.56331surveyors 57.67687In-sample accuracy and out of sample accuracy
# test MSEtest %>%add_predictions(model4) %>%summarise(MSE = mean((prestige - pred)^2, na.rm=TRUE)) MSE1 41.17247# training MSEtrain %>%add_predictions(model4) %>%summarise(MSE = mean((prestige - pred)^2, na.rm=TRUE)) MSE1 42.2731Out of sample accuracy: model 1, model 2 and model 3
# test MSEtest %>%add_predictions(model1) %>%summarise(MSE = mean((prestige - pred)^2, na.rm=TRUE)) MSE1 104.0138# test MSEtest %>%add_predictions(model2) %>%summarise(MSE = mean((prestige - pred)^2, na.rm=TRUE)) MSE1 69.17519# test MSEtest %>%add_predictions(model3) %>%summarise(MSE = mean((prestige - pred)^2, na.rm=TRUE)) MSE1 43.92756Model 4: 42.51
Modelling cycle
EDA
Fit
Examine the residuals (multicollinearity/ Influential cases)
Transform/ Add/ Drop regressors if necessary
Repeat above until you find a good model(s)
Use out-of-sample accuracy to select the final model
Presenting results
EDA
Final regression fit:
- sample size
- estimated coefficients and standard errors
- R2adj
- Visualizations (model fit, coefficients and CIs)
Model adequacy checking results: Residual plots and interpretations
Hypothesis testing and interpretations
- ANOVA, etc.
Out-of sample accuracy
- Some flexibility is possible in the presentation of results and you may want to adjust the rules above to emphasize the point you are trying to make.
Other models
Decision trees
Random forests
XGBoost
Deep learning approaches and many more..