STA 326 2.0 Programming and Data Analysis with R
Regression Analysis
Dr Thiyanga Talagala
Online distance learning/teaching materials during the COVID-19 outbreak.
Packages
library(broom)library(modelr)library(GGally)library(carData)library(tidyverse)library(magrittr)library(car) # to calculate VIFData: Prestige of Canadian Occupations
head(Prestige, 5) education income women prestige census typegov.administrators 13.11 12351 11.16 68.8 1113 profgeneral.managers 12.26 25879 4.02 69.1 1130 profaccountants 12.77 9271 15.70 63.4 1171 profpurchasing.officers 11.42 8865 9.11 56.8 1175 profchemists 14.62 8403 11.68 73.5 2111 profsummary(Prestige) education income women prestige Min. : 6.380 Min. : 611 Min. : 0.000 Min. :14.80 1st Qu.: 8.445 1st Qu.: 4106 1st Qu.: 3.592 1st Qu.:35.23 Median :10.540 Median : 5930 Median :13.600 Median :43.60 Mean :10.738 Mean : 6798 Mean :28.979 Mean :46.83 3rd Qu.:12.648 3rd Qu.: 8187 3rd Qu.:52.203 3rd Qu.:59.27 Max. :15.970 Max. :25879 Max. :97.510 Max. :87.20 census type Min. :1113 bc :44 1st Qu.:3120 prof:31 Median :5135 wc :23 Mean :5402 NA's: 4 3rd Qu.:8312 Max. :9517Data description
prestige: prestige of Canadian occupations, measured by the Pineo-Porter prestige score for occupation taken from a social survey conducted in the mid-1960s.
education: Average education of occupational incumbents, years, in 1971.
income: Average income of incumbents, dollars, in 1971.
women: Percentage of incumbents who are women.
census: Canadian Census occupational code.
type: Type of occupation.
- prof: professional and technical- wc: white collar- bc: blue collar- NA: missingThe dataset consists of 102 observations, each corresponding to a particular occupation.
Training test and Test set
smp_size <- 80## set the seed to make your partition reproducibleset.seed(123)train_ind <- sample(seq_len(nrow(Prestige)), size = smp_size)train <- Prestige[train_ind, ]dim(train)[1] 80 6test <- Prestige[-train_ind, ]dim(test)[1] 22 6Exploratory Data Analysis
Prestige_1 <- train %>% pivot_longer(c(1, 2, 3, 4), names_to="variable", values_to="value")Prestige_1# A tibble: 320 x 4 census type variable value <int> <fct> <chr> <dbl> 1 3156 wc education 12.8 2 3156 wc income 5180 3 3156 wc women 76.0 4 3156 wc prestige 67.5 5 8335 bc education 7.92 6 8335 bc income 6477 7 8335 bc women 5.17 8 8335 bc prestige 41.8 9 5133 wc education 11.1 10 5133 wc income 8780 # … with 310 more rowsExploratory Data Analysis
Prestige_1 <- train %>% pivot_longer(c(1, 2, 3, 4), names_to="variable", values_to="value")Prestige_1# A tibble: 320 x 4 census type variable value <int> <fct> <chr> <dbl> 1 3156 wc education 12.8 2 3156 wc income 5180 3 3156 wc women 76.0 4 3156 wc prestige 67.5 5 8335 bc education 7.92 6 8335 bc income 6477 7 8335 bc women 5.17 8 8335 bc prestige 41.8 9 5133 wc education 11.1 10 5133 wc income 8780 # … with 310 more rowshead(train) education income women prestige census typemedical.technicians 12.79 5180 76.04 67.5 3156 wcwelders 7.92 6477 5.17 41.8 8335 bccommercial.travellers 11.13 8780 3.16 40.2 5133 wceconomists 14.44 8049 57.31 62.2 2311 proffarmers 6.84 3643 3.60 44.1 7112 <NA>receptionsts 11.04 2901 92.86 38.7 4171 wcExploratory Data Analysis
ggplot(Prestige_1, aes(x=value)) + geom_histogram() + facet_wrap(variable ~., ncol=1)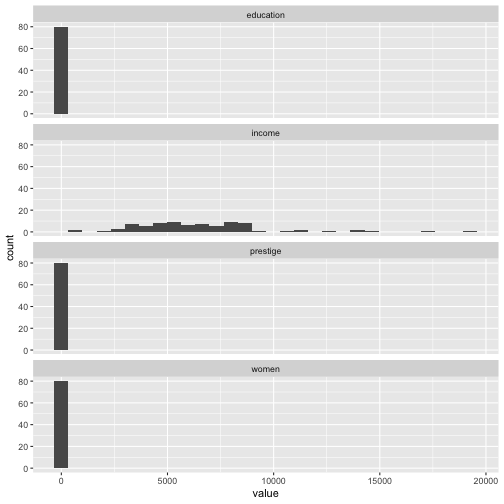
Exploratory Data Analysis
ggplot(Prestige_1, aes(x=value)) + geom_histogram() + facet_wrap(variable ~., ncol=1, scales = "free")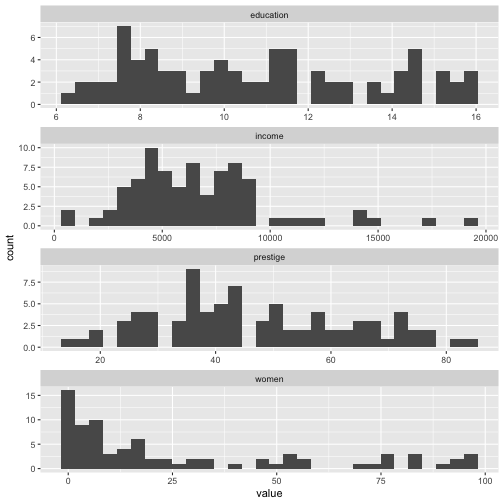
Exploratory Data Analysis
ggplot(Prestige_1, aes(x=value)) + geom_histogram(colour="white") + facet_wrap(variable ~., ncol=1, scales = "free")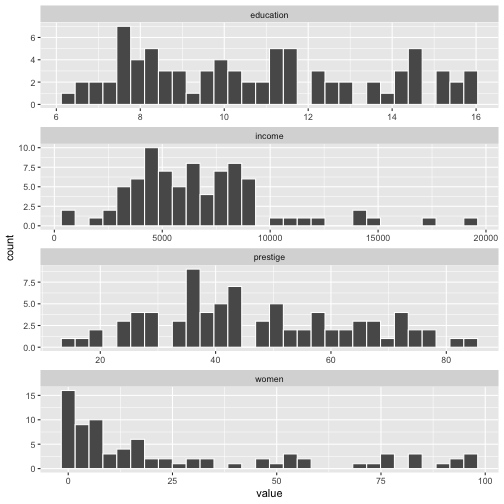
Exploratory Data Analysis
ggplot(Prestige_1, aes(x=value, fill=variable)) + geom_density() + facet_wrap(variable ~., ncol=1, scales = "free")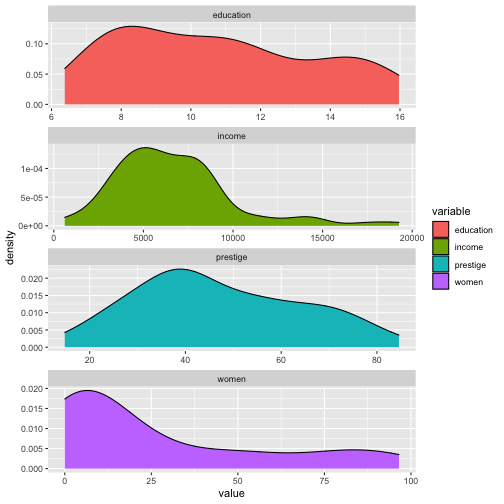
Exploratory Data Analysis
ggplot(Prestige_1, aes(y = value, x = type, fill = type)) + geom_boxplot() + facet_wrap(variable ~., ncol=1, scales = "free")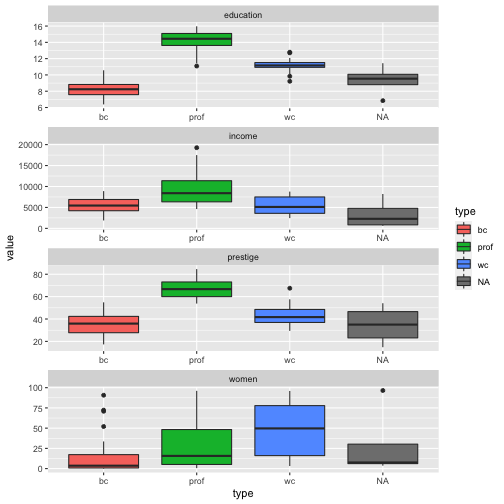
Exploratory Data Analysis
Prestige_1 %>% filter(is.na(type) == FALSE) %>%ggplot(aes(y=value, x=type, fill=type)) + geom_boxplot() + facet_wrap(variable ~., ncol=1, scales = "free")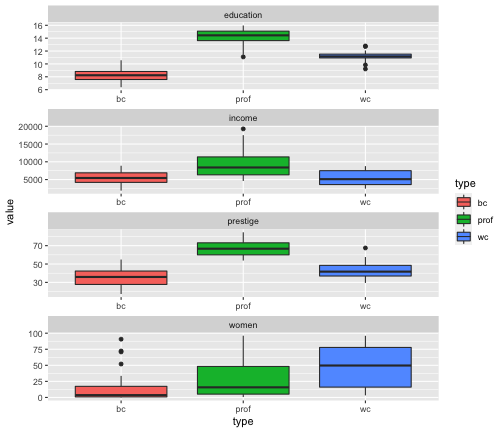
Exploratory Data Analysis
Prestige_1 %>% filter(is.na(type) == FALSE) %>%ggplot(aes(x = value, y = type, fill = type)) + geom_boxplot() + facet_wrap(variable ~., ncol=1, scales = "free")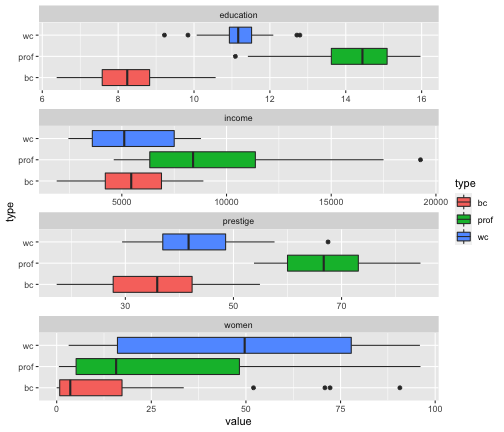
Exploratory Data Analysis
train %>% select(education, income, prestige, women) %>% ggpairs()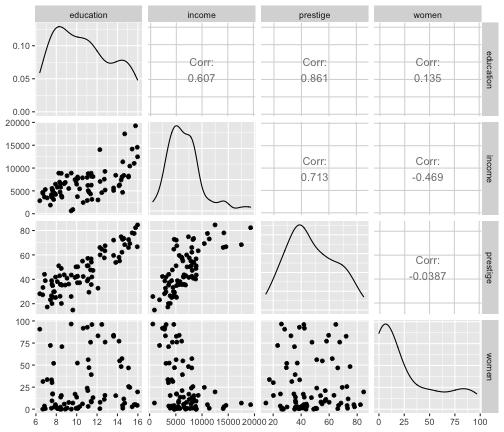
Exploratory Data Analysis
train %>% filter(is.na(type) == FALSE) %>% ggpairs(columns= c("education", "income", "prestige", "women"), mapping=aes(color=type))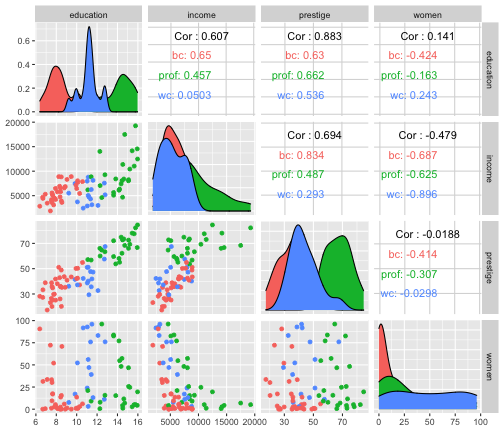
Regression analysis
Steps
Fit a model.
Visualize the fitted model.
Measuring the strength of the fit.
Residual analysis.
Interpret the coefficients.
Make predictions using the fitted model.
Model 1: prestige ~ education
Recap
True relationship between X and Y in the population
Y=f(X)+ϵ
If f is approximated by a linear function
Y=β0+β1X+ϵ
The error terms are normally distributed with mean 0 and variance σ2. Then the mean response, Y, at any value of the X is
E(Y|X=xi)=E(β0+β1xi+ϵ)=β0+β1xi
For a single unit (yi,xi)
yi=β0+β1xi+ϵi where ϵi∼N(0,σ2)
We use sample values (yi,xi) where i=1,2,...n to estimate β0 and β1.
The fitted regression model is
^Yi=^β0+^β1xi
How to estimate β0 and β1?
Sum of squares of Residuals
SSR=e21+e22+...+e2n
The least-squares regression approach chooses coefficients ^β0 and ^β1 to minimize SSR.
Model 1: prestige ~ education
1. Fit a model
Model 1: Fit a model
yi=β0+β1xi+ϵi,where ϵi∼NID(0,σ2)
To estimate β0 and β1
model1 <- lm(prestige ~ education, data=train)summary(model1)Call:lm(formula = prestige ~ education, data = train)Residuals: Min 1Q Median 3Q Max -26.4715 -5.8396 0.9778 6.2464 17.4767 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) -9.418 3.918 -2.404 0.0186 * education 5.269 0.353 14.928 <2e-16 ***---Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Residual standard error: 8.754 on 78 degrees of freedomMultiple R-squared: 0.7407, Adjusted R-squared: 0.7374 F-statistic: 222.8 on 1 and 78 DF, p-value: < 2.2e-16What's messy about the output?
Call:lm(formula = prestige ~ education, data = train)Residuals: Min 1Q Median 3Q Max -26.4715 -5.8396 0.9778 6.2464 17.4767 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) -9.418 3.918 -2.404 0.0186 * education 5.269 0.353 14.928 <2e-16 ***---Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Residual standard error: 8.754 on 78 degrees of freedomMultiple R-squared: 0.7407, Adjusted R-squared: 0.7374 F-statistic: 222.8 on 1 and 78 DF, p-value: < 2.2e-16Extract coefficients takes multiple steps.
data.frame(coef(summary(model1)))Column names are inconvenient to use.
Information are stored in row names.
broom functions
broom takes model objects and turns them into tidy data frames that can be used with other tidy tools.
Three main functions
tidy(): component-level statisticsaugment(): observation-level statisticsglance(): model-level statistics
Component-level statistics: tidy()
model1 %>% tidy()# A tibble: 2 x 5 term estimate std.error statistic p.value <chr> <dbl> <dbl> <dbl> <dbl>1 (Intercept) -9.42 3.92 -2.40 1.86e- 22 education 5.27 0.353 14.9 1.43e-24model1 %>% tidy(conf.int=TRUE)# A tibble: 2 x 7 term estimate std.error statistic p.value conf.low conf.high <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>1 (Intercept) -9.42 3.92 -2.40 1.86e- 2 -17.2 -1.622 education 5.27 0.353 14.9 1.43e-24 4.57 5.97model1 %>% tidy() %>% select(term, estimate)# A tibble: 2 x 2 term estimate <chr> <dbl>1 (Intercept) -9.422 education 5.27Component-level statistics: tidy() (cont.)
model1 %>% tidy()# A tibble: 2 x 5 term estimate std.error statistic p.value <chr> <dbl> <dbl> <dbl> <dbl>1 (Intercept) -9.42 3.92 -2.40 1.86e- 22 education 5.27 0.353 14.9 1.43e-24Fitted model is
^Yi=−9.42+5.27xi
Why are tidy model outputs useful?
tidy_model1 <- model1 %>% tidy(conf.int=TRUE)ggplot(tidy_model1, aes(x=estimate, y=term, color=term)) + geom_point() + geom_errorbarh(aes(xmin = conf.low, xmax=conf.high))+ggtitle("Coefficient plot")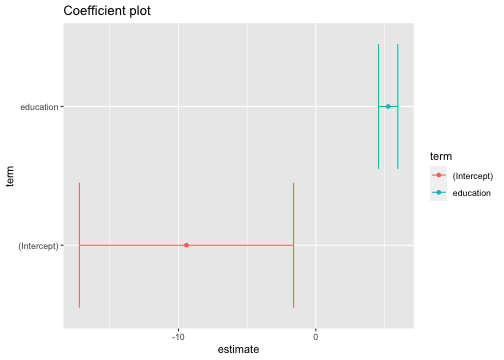
Model 1: prestige ~ education
1. Fit a model
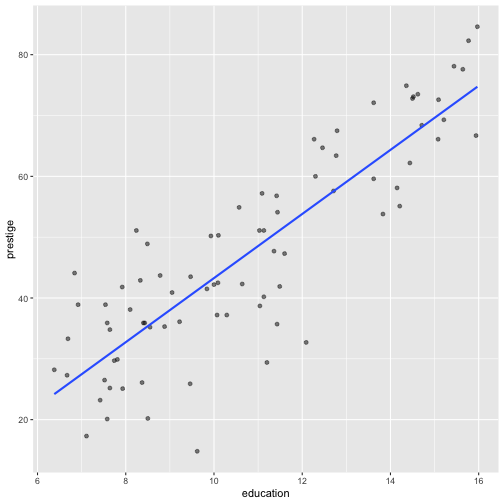
2. Visualise the fitted model
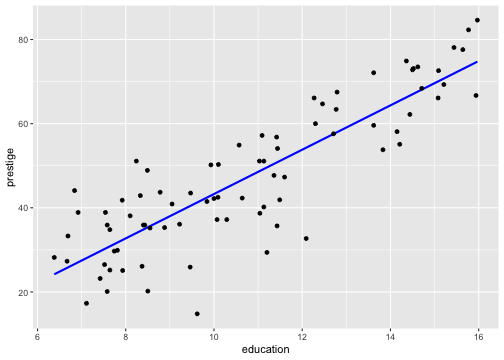
Model 1: Visualise the fitted model
Model 1: Visualise the fitted model (style the line)
ggplot(data=train, aes(y=prestige, x=education)) + geom_point(alpha=0.5) + geom_smooth(method="lm", se=FALSE, col="forestgreen", lwd=2)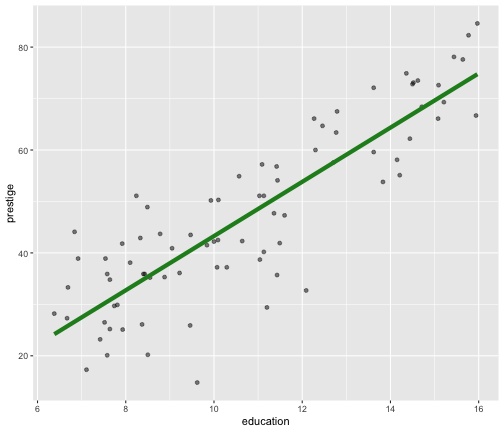
Model 1: prestige ~ education
1. Fit a model
2. Visualise the fitted model
3. Measure the strength of the fit
Model-level statistics: glance()
Measuring the strength of the fit
glance(model1)# A tibble: 1 x 11 r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC <dbl> <dbl> <dbl> <dbl> <dbl> <int> <dbl> <dbl> <dbl>1 0.741 0.737 8.75 223. 1.43e-24 2 -286. 578. 585.# … with 2 more variables: deviance <dbl>, df.residual <int>glance(model1)$adj.r.squared # extract values[1] 0.7374038Roughly 73% of the variability in prestige can be explained by the variable education.
Model 1: prestige ~ education
1. Fit a model
2. Visualise the fitted model
3. Measure the strength of the fit
4. Residual analysis
Observation-level statistics: augment()
model1_fitresid <- augment(model1)model1_fitresid# A tibble: 80 x 10 .rownames prestige education .fitted .se.fit .resid .hat .sigma .cooksd <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> 1 medical.… 67.5 12.8 58.0 1.22 9.53 0.0193 8.74 1.19e-2 2 welders 41.8 7.92 32.3 1.40 9.49 0.0255 8.74 1.58e-2 3 commerci… 40.2 11.1 49.2 0.988 -9.03 0.0127 8.75 6.95e-3 4 economis… 62.2 14.4 66.7 1.63 -4.47 0.0347 8.80 4.85e-3 5 farmers 44.1 6.84 26.6 1.69 17.5 0.0373 8.57 8.03e-2 6 receptio… 38.7 11.0 48.8 0.984 -10.1 0.0126 8.74 8.55e-3 7 sales.su… 41.5 9.84 42.4 1.03 -0.931 0.0138 8.81 8.04e-5 8 mail.car… 36.1 9.22 39.2 1.12 -3.06 0.0163 8.80 1.03e-3 9 taxi.dri… 25.1 7.93 32.4 1.40 -7.27 0.0254 8.77 9.22e-310 veterina… 66.7 15.9 74.6 2.08 -7.87 0.0563 8.76 2.56e-2# … with 70 more rows, and 1 more variable: .std.resid <dbl>Residuals and Fitted Values
Residuals and Fitted Values
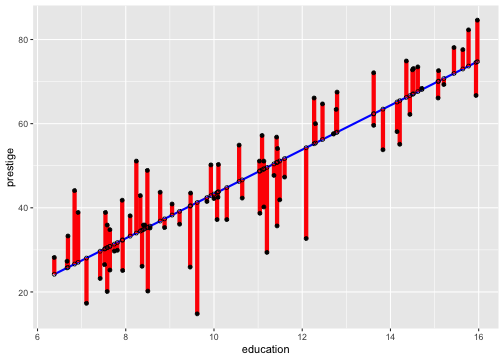
The residual is the difference between the observed and predicted response.
The residual for the ith observation is
ei=yi−^Yi=yi−(^β0+^β1xi)
Conditions for inference for regression
The relationship between the response and the predictors is linear.
The error terms are assumed to have zero mean and unknown constant variance σ2.
The errors are normally distributed.
The errors are uncorrelated.
Plot of residuals in time sequence.
The errors are uncorrelated.
Often, we can conclude that the this assumption is sufficiently met based on a description of the data and how it was collected.
Plot of residuals vs fitted values
This is useful for detecting several common types of model inadequacies.
Plot of residuals vs fitted values and Plot of residuals vs predictor
linearity and constant variance
Residuals vs Fitted
ggplot(model1_fitresid, aes(x = .fitted, y = .resid))+ ------ + ------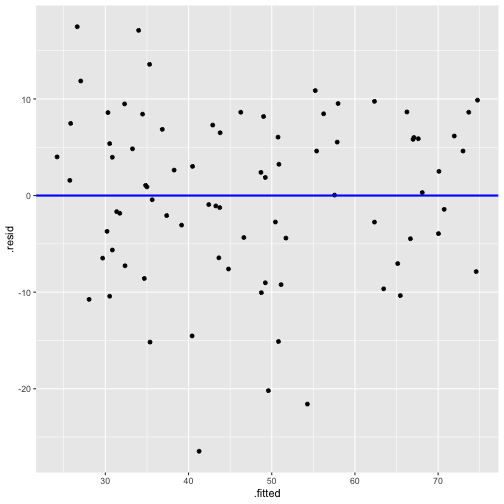
Residuals vs X
ggplot(model1_fitresid, aes(x = education, y = .resid))+ ------ + ------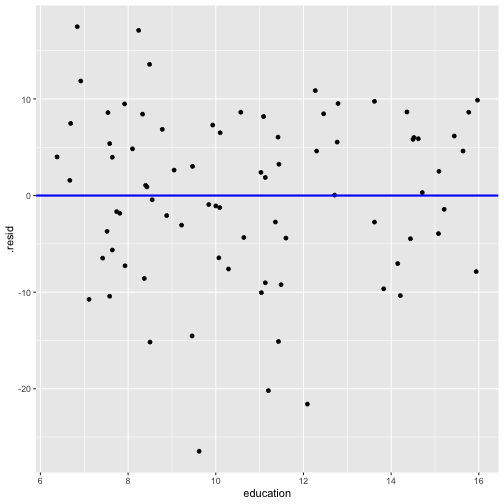
Normality of residuals
ggplot(model1_fitresid, aes(x=.resid))+ geom_histogram(colour="white")+ggtitle("Distribution of Residuals")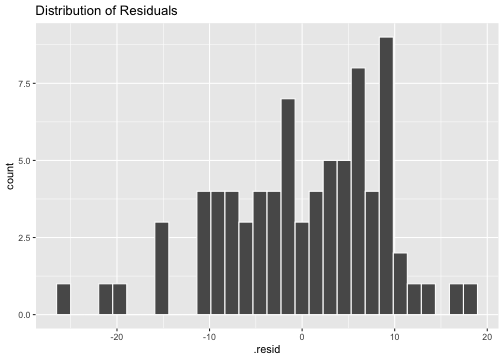
ggplot(model1_fitresid, aes(sample=.resid))+ stat_qq() + stat_qq_line()+labs(x="Theoretical Quantiles", y="Sample Quantiles")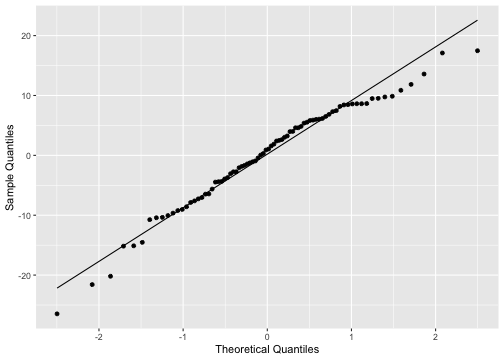
shapiro.test(model1_fitresid$.resid) Shapiro-Wilk normality testdata: model1_fitresid$.residW = 0.97192, p-value = 0.07527Model 2: prestige ~ education + income
1. Fit a model
2. Visualise the fitted model
3. Measure the strength of the fit
4. Residual analysis
Model 2: prestige ~ education + income
model2 <- lm(prestige ~ income + education, data=train)summary(model2)Call:lm(formula = prestige ~ income + education, data = train)Residuals: Min 1Q Median 3Q Max -18.9407 -4.1745 0.2026 4.9471 17.7176 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) -7.5400414 3.4980062 -2.156 0.0342 * income 0.0015286 0.0003249 4.705 1.1e-05 ***education 4.1452613 0.3937971 10.526 < 2e-16 ***---Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Residual standard error: 7.765 on 77 degrees of freedomMultiple R-squared: 0.7986, Adjusted R-squared: 0.7934 F-statistic: 152.7 on 2 and 77 DF, p-value: < 2.2e-16Model 2: prestige ~ education + income (cont.)
model2_fitresid <- augment(model2)model2_fitresid# A tibble: 80 x 11 .rownames prestige income education .fitted .se.fit .resid .hat .sigma <chr> <dbl> <int> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> 1 medical.… 67.5 5180 12.8 53.4 1.45 14.1 0.0350 7.64 2 welders 41.8 6477 7.92 35.2 1.38 6.61 0.0317 7.78 3 commerci… 40.2 8780 11.1 52.0 1.06 -11.8 0.0186 7.70 4 economis… 62.2 8049 14.4 64.6 1.51 -2.42 0.0378 7.81 5 farmers 44.1 3643 6.84 26.4 1.50 17.7 0.0374 7.54 6 receptio… 38.7 2901 11.0 42.7 1.56 -3.96 0.0405 7.80 7 sales.su… 41.5 7482 9.84 44.7 1.03 -3.19 0.0177 7.81 8 mail.car… 36.1 5511 9.22 39.1 0.991 -3.00 0.0163 7.81 9 taxi.dri… 25.1 4224 7.93 31.8 1.24 -6.69 0.0257 7.7810 veterina… 66.7 14558 15.9 80.8 2.27 -14.1 0.0853 7.63# … with 70 more rows, and 2 more variables: .cooksd <dbl>, .std.resid <dbl>Plot of residuals vs fitted values
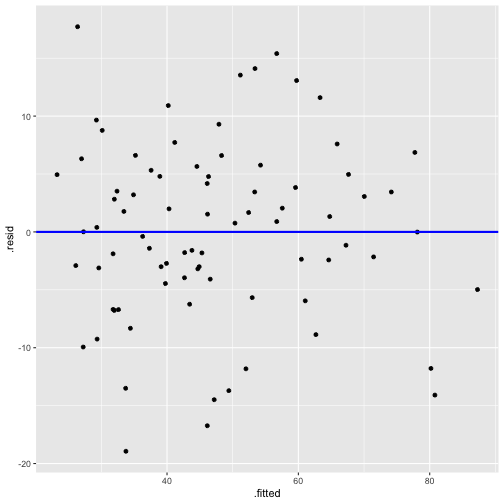
linearity and constant variance?
Normality of residuals
ggplot(model2_fitresid, aes(x=.resid))+ geom_histogram(colour="white")+ggtitle("Distribution of Residuals")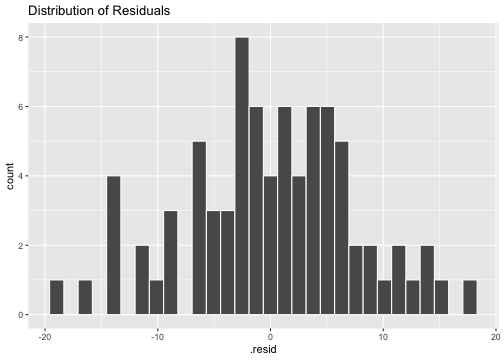
ggplot(model2_fitresid, aes(sample=.resid))+ stat_qq() + stat_qq_line()+labs(x="Theoretical Quantiles", y="Sample Quantiles")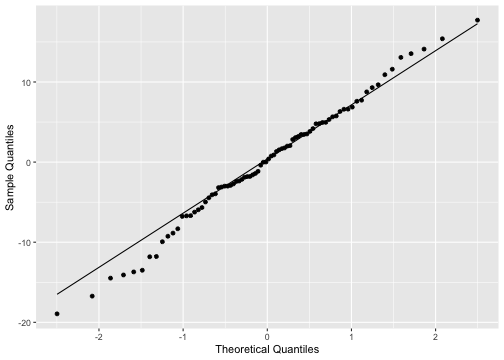
shapiro.test(model2_fitresid$.resid) Shapiro-Wilk normality testdata: model2_fitresid$.residW = 0.99183, p-value = 0.8977Plot of residuals vs predictor variables
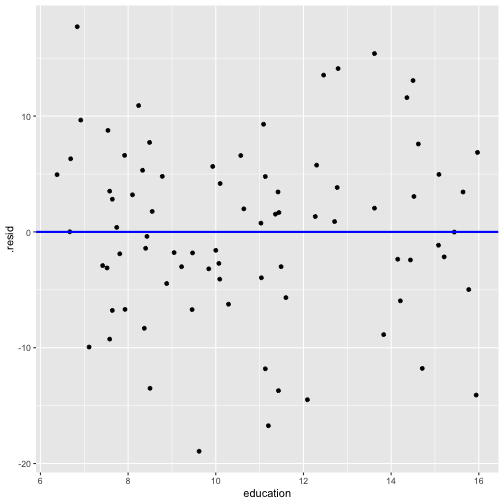
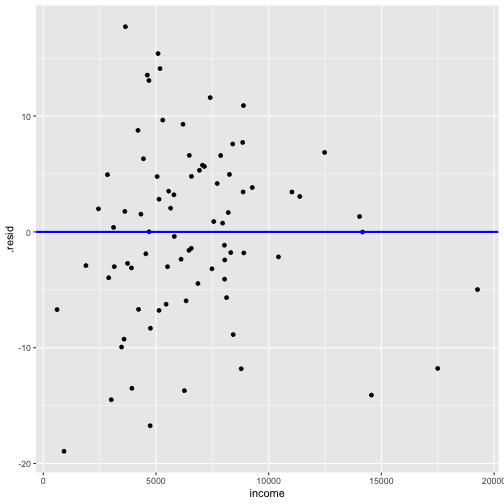
Model 3: prestige ~ education + log(income)
1. Fit a model
2. Visualise the fitted model
3. Measure the strength of the fit
4. Residual analysis
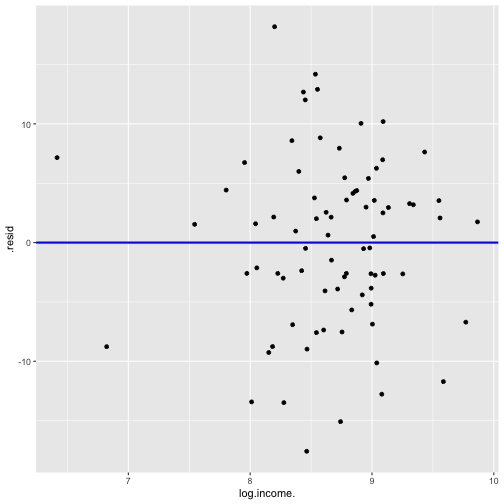
Model 3: prestige ~ education + log(income)
model3 <- lm(prestige ~ log(income) + education, data=train)summary(model3)Call:lm(formula = prestige ~ log(income) + education, data = train)Residuals: Min 1Q Median 3Q Max -17.5828 -4.1534 0.7978 4.3347 18.1881 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) -86.5942 13.4064 -6.459 8.62e-09 ***log(income) 10.2025 1.7189 5.936 7.91e-08 ***education 4.2163 0.3436 12.271 < 2e-16 ***---Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Residual standard error: 7.298 on 77 degrees of freedomMultiple R-squared: 0.8221, Adjusted R-squared: 0.8175 F-statistic: 177.9 on 2 and 77 DF, p-value: < 2.2e-16model3_fitresid <- augment(model3)Plot of Residuals vs Fitted
Now - Model 3
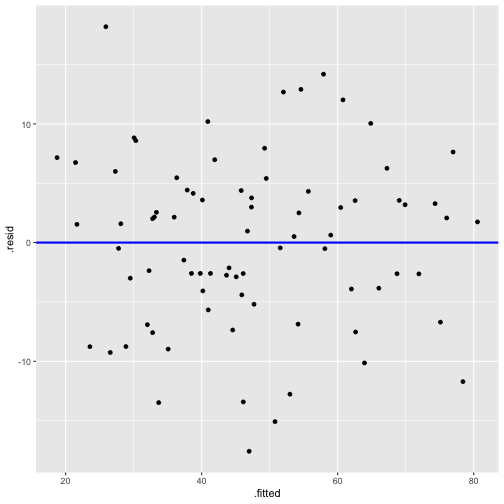
Before - Model 2
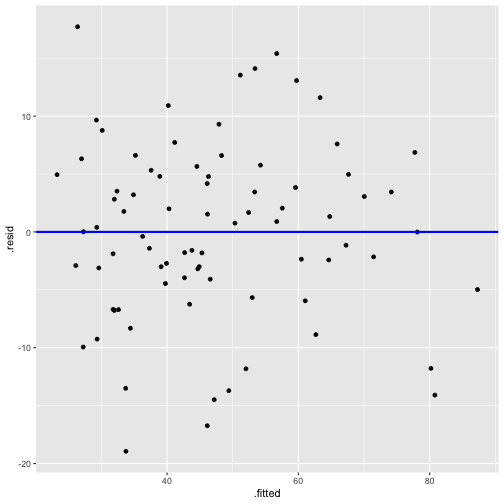
Normality of residuals
ggplot(model3_fitresid, aes(x=.resid))+ geom_histogram(colour="white")+ggtitle("Distribution of Residuals")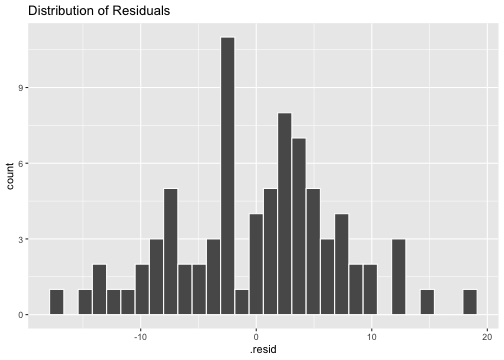
ggplot(model3_fitresid, aes(sample=.resid))+ stat_qq() + stat_qq_line()+labs(x="Theoretical Quantiles", y="Sample Quantiles")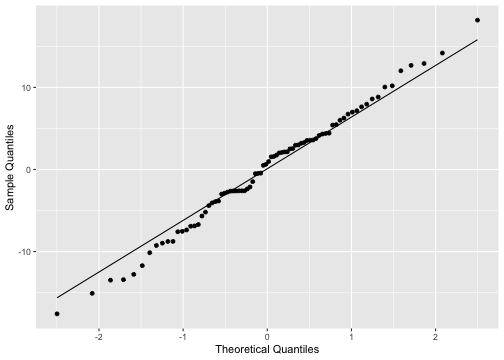
shapiro.test(model3_fitresid$.resid) Shapiro-Wilk normality testdata: model3_fitresid$.residW = 0.9931, p-value = 0.9492Plot of residuals vs predictor variables
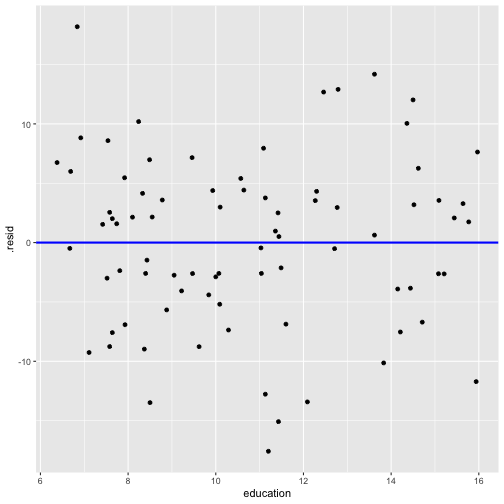
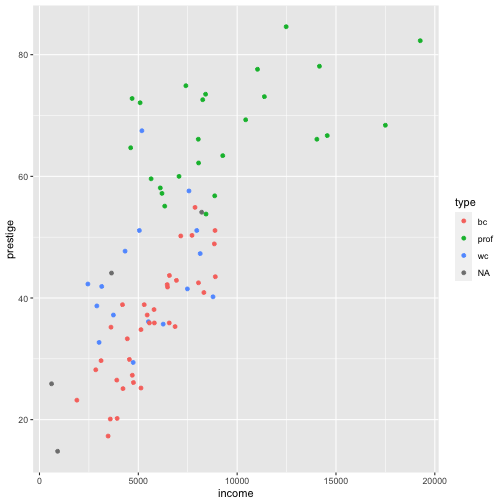
Prestige vs income by type
R code: ___
Prestige vs income by type
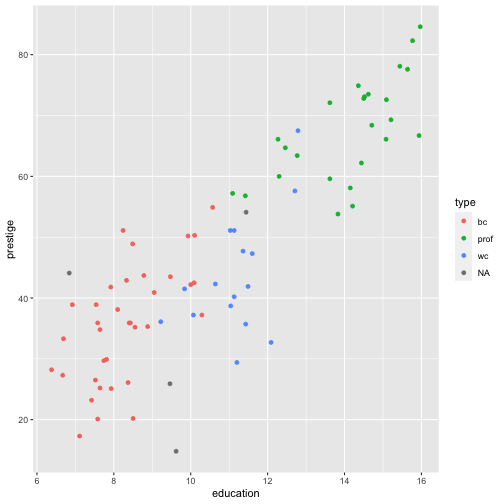
R code: __
Model 4: prestige ~ education + log(income) + type
1. Fit a model
2. Visualise the fitted model
3. Measure the strength of the fit
4. Residual analysis
str(train)'data.frame': 80 obs. of 6 variables: $ education: num 12.79 7.92 11.13 14.44 6.84 ... $ income : int 5180 6477 8780 8049 3643 2901 7482 5511 4224 14558 ... $ women : num 76.04 5.17 3.16 57.31 3.6 ... $ prestige : num 67.5 41.8 40.2 62.2 44.1 38.7 41.5 36.1 25.1 66.7 ... $ census : int 3156 8335 5133 2311 7112 4171 5130 4172 9173 3115 ... $ type : Factor w/ 3 levels "bc","prof","wc": 3 1 3 2 NA 3 3 3 1 2 ...Model 4: prestige ~ education + log(income) + type
model4 <- lm(prestige ~ log(income) + education + type, data=train)summary(model4)Call:lm(formula = prestige ~ log(income) + education + type, data = train)Residuals: Min 1Q Median 3Q Max -13.8420 -3.7790 0.6321 4.7393 17.8611 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) -71.9102 17.7934 -4.041 0.000133 ***log(income) 9.1505 2.2945 3.988 0.000160 ***education 3.5136 0.7553 4.652 1.48e-05 ***typeprof 6.7702 4.3797 1.546 0.126595 typewc -1.6497 3.0213 -0.546 0.586758 ---Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Residual standard error: 6.746 on 71 degrees of freedom (4 observations deleted due to missingness)Multiple R-squared: 0.8494, Adjusted R-squared: 0.8409 F-statistic: 100.1 on 4 and 71 DF, p-value: < 2.2e-16Model 4: prestige ~ education + log(income) + type
model4_fitresid <- augment(model4)head(model4_fitresid)# A tibble: 6 x 12 .rownames prestige log.income. education type .fitted .se.fit .resid .hat <chr> <dbl> <dbl> <dbl> <fct> <dbl> <dbl> <dbl> <dbl>1 medical.… 67.5 8.55 12.8 wc 49.6 2.06 17.9 0.09342 welders 41.8 8.78 7.92 bc 36.2 1.31 5.58 0.03773 commerci… 40.2 9.08 11.1 wc 48.6 2.14 -8.43 0.100 4 economis… 62.2 8.99 14.4 prof 67.9 1.40 -5.69 0.04315 receptio… 38.7 7.97 11.0 wc 38.2 2.07 0.515 0.09446 sales.su… 41.5 8.92 9.84 wc 42.6 2.36 -1.14 0.122 # … with 3 more variables: .sigma <dbl>, .cooksd <dbl>, .std.resid <dbl>Plot of Residuals vs Fitted
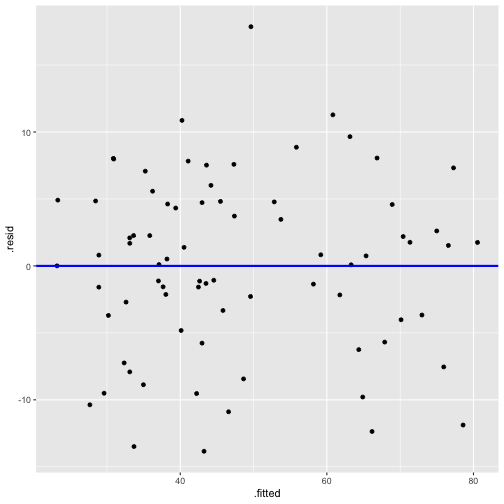
Normality of residuals
ggplot(model4_fitresid, aes(x=.resid))+ geom_histogram(colour="white")+ggtitle("Distribution of Residuals")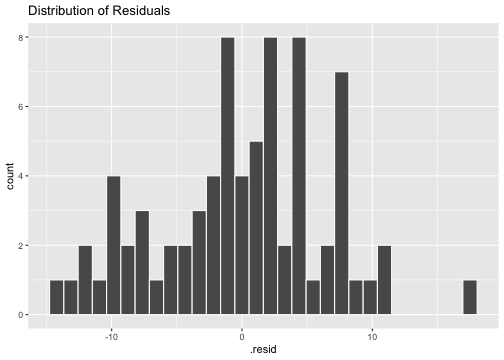
ggplot(model4_fitresid, aes(sample=.resid))+ stat_qq() + stat_qq_line()+labs(x="Theoretical Quantiles", y="Sample Quantiles")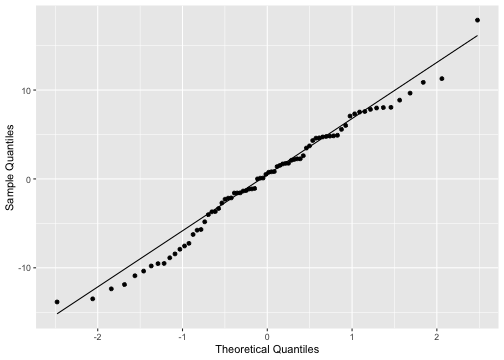
shapiro.test(model4_fitresid$.resid) Shapiro-Wilk normality testdata: model4_fitresid$.residW = 0.9838, p-value = 0.4445Plot of residuals vs predictor variables
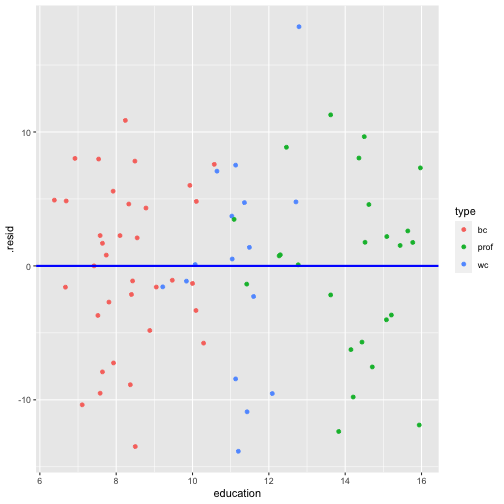
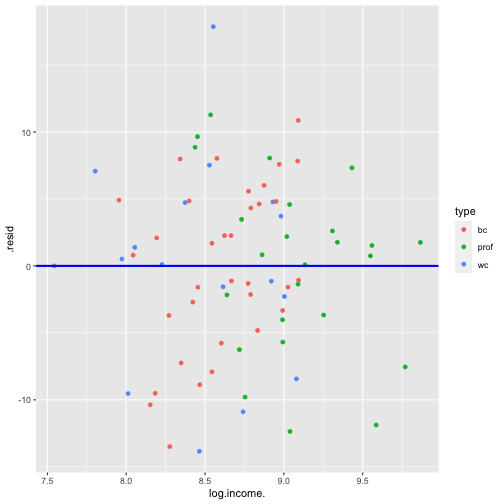
Multicollinearity
car::vif(model4) GVIF Df GVIF^(1/(2*Df))log(income) 1.789890 1 1.337868education 7.471481 1 2.733401type 7.021948 2 1.627850VIFs larger than 10 imply series problems with multicollinearity.
Detecting influential observations
library(lindia)gg_cooksd(model4)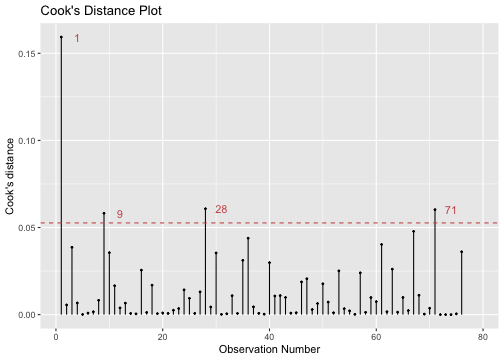
Influential outliers (cont.)
model4_fitresid %>% top_n(4, wt = .cooksd)# A tibble: 4 x 12 .rownames prestige log.income. education type .fitted .se.fit .resid .hat <chr> <dbl> <dbl> <dbl> <fct> <dbl> <dbl> <dbl> <dbl>1 medical.… 67.5 8.55 12.8 wc 49.6 2.06 17.9 0.09342 veterina… 66.7 9.59 15.9 prof 78.6 1.90 -11.9 0.07953 file.cle… 32.7 8.01 12.1 wc 42.2 2.32 -9.53 0.118 4 collecto… 29.4 8.46 11.2 wc 43.2 1.69 -13.8 0.0629# … with 3 more variables: .sigma <dbl>, .cooksd <dbl>, .std.resid <dbl>Solutions
Remove influential observations, and re-fit model.
Transform explanatory variables to reduce influence.
Use weighted regression to down weight influence of extreme observations.
Hypothesis testing
Y=β0+β1x1+β2x2+β3x3+β4x4+ϵ
What is the overall adequacy of the model?
H0:β1=β2=β3=β4
H1:βj≠0 for at least one j,j=1,2,3,4
Which specific regressors seem important?
H0:β1=0
H1:β1≠0
Making predictions
Method 1
head(test) education income women prestige census typegov.administrators 13.11 12351 11.16 68.8 1113 profgeneral.managers 12.26 25879 4.02 69.1 1130 profmining.engineers 14.64 11023 0.94 68.8 2153 profsurveyors 12.39 5902 1.91 62.0 2161 profvocational.counsellors 15.22 9593 34.89 58.3 2391 profphysicians 15.96 25308 10.56 87.2 3111 profpredict(model4, test) gov.administrators general.managers mining.engineers 67.13432 70.91638 71.46917 surveyors vocational.counsellors physicians 57.84739 72.23557 83.71240 nursing.aides secretaries bookkeepers 35.92664 43.13913 42.87183 shipping.clerks telephone.operators office.clerks 36.14801 37.10841 41.15412 sales.clerks service.station.attendant real.estate.salesmen 33.68323 34.08491 46.41067 policemen launderers farm.workers 49.69682 27.10663 26.13155 textile.labourers machinists electronic.workers 26.40488 39.63996 34.62981 masons 30.82164Making predictions
Method 2
library(modelr)test <- test %>% add_predictions(model4)test education income women prestige census type predgov.administrators 13.11 12351 11.16 68.8 1113 prof 67.13432general.managers 12.26 25879 4.02 69.1 1130 prof 70.91638mining.engineers 14.64 11023 0.94 68.8 2153 prof 71.46917surveyors 12.39 5902 1.91 62.0 2161 prof 57.84739vocational.counsellors 15.22 9593 34.89 58.3 2391 prof 72.23557physicians 15.96 25308 10.56 87.2 3111 prof 83.71240nursing.aides 9.45 3485 76.14 34.9 3135 bc 35.92664secretaries 11.59 4036 97.51 46.0 4111 wc 43.13913bookkeepers 11.32 4348 68.24 49.4 4131 wc 42.87183shipping.clerks 9.17 4761 11.37 30.9 4153 wc 36.14801telephone.operators 10.51 3161 96.14 38.1 4175 wc 37.10841office.clerks 11.00 4075 63.23 35.6 4197 wc 41.15412sales.clerks 10.05 2594 67.82 26.5 5137 wc 33.68323service.station.attendant 9.93 2370 3.69 23.3 5145 bc 34.08491real.estate.salesmen 11.09 6992 24.44 47.1 5172 wc 46.41067policemen 10.93 8891 1.65 51.6 6112 bc 49.69682launderers 7.33 3000 69.31 20.8 6162 bc 27.10663farm.workers 8.60 1656 27.75 21.5 7182 bc 26.13155textile.labourers 6.74 3485 39.48 28.8 8278 bc 26.40488machinists 8.81 6686 4.28 44.2 8313 bc 39.63996electronic.workers 8.76 3942 74.54 50.8 8534 bc 34.62981masons 6.60 5959 0.52 36.2 8782 bc 30.82164In-sample accuracy and out of sample accuracy
# test MSEtest %>%add_predictions(model4) %>%summarise(MSE = mean((prestige - pred)^2, na.rm=TRUE)) MSE1 40.82736# training MSEtrain %>%add_predictions(model4) %>%summarise(MSE = mean((prestige - pred)^2, na.rm=TRUE)) MSE1 42.51919Out of sample accuracy: model 1, model 2 and model 3
# test MSEtest %>%add_predictions(model1) %>%summarise(MSE = mean((prestige - pred)^2, na.rm=TRUE)) MSE1 105.7605# test MSEtest %>%add_predictions(model2) %>%summarise(MSE = mean((prestige - pred)^2, na.rm=TRUE)) MSE1 67.16647# test MSEtest %>%add_predictions(model3) %>%summarise(MSE = mean((prestige - pred)^2, na.rm=TRUE)) MSE1 45.36052Model 4: 42.51
Modelling cycle
EDA
Fit
Examine the residuals (multicollinearity/ Influential cases)
Transform/ Add/ Drop regressors if necessary
Repeat above until you find a good model(s)
Use out-of-sample accuracy to select the final model
Presenting results
EDA
Final regression fit:
- sample size
- estimated coefficients and standard errors
- R2adj
- Visualizations (model fit, coefficients and CIs)
Model adequacy checking results: Residual plots and interpretations
Hypothesis testing and interpretations
- ANOVA, etc.
Out-of sample accuracy
- Some flexibility is possible in the presentation of results and you may want to adjust the rules above to emphasize the point you are trying to make.
Other models
Decision trees
Random forests
XGBoost
Deep learning approaches and many more..