STA 326 2.0 Programming and Data Analysis with R
🎉 Introduction to R, RStudio and R Programming Basics
Dr Thiyanga Talagala
Today's menu
What is R?
Why R?
R and Rstudio.
Installing R and Rstudio?
Familiarize with RStudio interface.
R Studio cloud.
Using R as a calculator.
Basic vector operations.

What statistical software packages are you familiar with?
02:30
R Programming Language
R is a software environment for statistical computing and graphics.
Language designers: Ross Ihaka and Robert Gentleman at the University of Auckland, New Zealand.
Parent language: S
R is a functional programming language. However, R system includes some support for object-oriented programming.

Minitab - Menu driven software
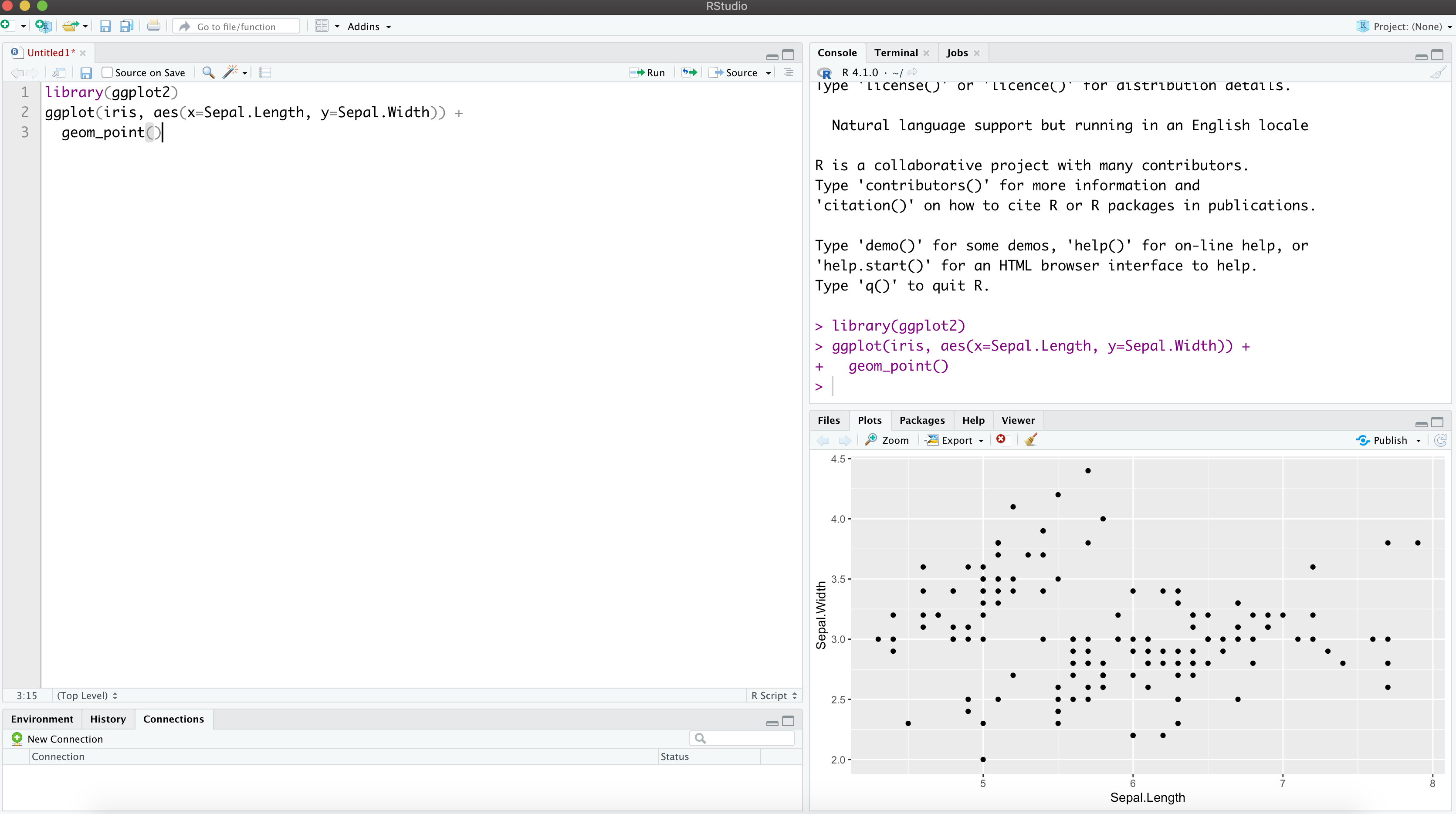
R - Scripting language
Menu driven software

Pros
Do not have to remember the commands.
User friendly.
Cons
Irritating if there are too many levels of menues to move around.
Difficult to reproduce results.
Scripting language

Pros
Useful for collaborative research.
Ideal for reproducible research
Cons
- The learning curve may be difficult at the start.
Why learn R?
Free and open-source software package
A large online community that makes it fun to learn
Latest cutting edge technology
Easier to update analysis
Easier to reproduce analysis
Easier to collaborate with others
Easier to automate analysis
R
R environment - macOS
R environment - macOS
R environment
The RStudio IDE
The RStudio IDE
R and RStudio
R and RStudio

If R were an airplane, RStudio would be the airport, providing many, many supporting services that make it easier for you, the pilot, to take off and go to awesome places. Sure, you can fly an airplane without an airport, but having those runways and supporting infrastructure is a game-changer."
Julie Lowndes
Create a new project
Get Working Directory
Display the current working directory.
getwd()Set Working Directory
setwd(include_path_here)Basics of R programming
R Console
7+1[1] 8rnorm(10) [1] -0.52734986 -1.00880796 -0.51885818 -1.25822706 -2.03810299 0.86445152 [7] -0.32695804 -0.27729466 -0.08115242 2.29875891Variable assignment
a <- rnorm(10)a [1] -0.17080946 -1.62197463 -0.19588570 -0.49785571 -0.01480748 1.49439259 [7] 1.11982158 -0.16474605 0.44264127 -0.18153232Variable assignment
a <- rnorm(10)a [1] -0.17080946 -1.62197463 -0.19588570 -0.49785571 -0.01480748 1.49439259 [7] 1.11982158 -0.16474605 0.44264127 -0.18153232b <- a*100b [1] -17.080946 -162.197463 -19.588570 -49.785571 -1.480748 149.439259 [7] 111.982158 -16.474605 44.264127 -18.153232Variable assignment
a <- rnorm(10)a [1] -0.17080946 -1.62197463 -0.19588570 -0.49785571 -0.01480748 1.49439259 [7] 1.11982158 -0.16474605 0.44264127 -0.18153232b <- a*100b [1] -17.080946 -162.197463 -19.588570 -49.785571 -1.480748 149.439259 [7] 111.982158 -16.474605 44.264127 -18.153232c <- "corona"c[1] "corona"Case sensitivity
R is case sensitive. The following are all different.
coronaCoronaCORONAWorking directory
To check your current working directory
getwd()To change the working directory
setwd("/Users/thiyanga/sta326")Data permanency
All variables are kept in the workspace.
ls()can be used to display the names of the objects which are currently stored within R.The collection of objects currently stored is called the workspace.
ls()[1] "a" "b" "c"Remove objects
- To remove objects the function
rmis available
Remove specific objects: rm(x, y, z)
rm(a)ls()[1] "b" "c"Remove all objects: rm(list=ls())
rm(list=ls())ls()character(0)Close the project
At the end of an R session, if you click save: the objects are written to a file called .RData in the current directory, and the command lines used in the session are saved to a file called .Rhistory
When R is started at later time from the same directory.
When R is started at later time from the same directory it reloads the associated workspace and commands history.
When R is started at later time from the same directory it reloads the associated workspace and commands history.
Comment your code
- Each line of a comment should begin with the comment symbol and a single space: # .
rnorm(10) # This is a comment [1] 1.1100805 -0.2128508 -0.3968625 1.4603542 1.4615554 0.2802397 [7] 0.7230058 -0.4670261 -0.0622365 0.9424283sum(1:10) # Summation of numbers from 1 to 10.[1] 55Style Guide
- Good coding style is like correct punctuation: you can manage without it, butitsuremakesthingseasiertoread. -- Hadley Wickham
sum(1:10)#Bad commenting style[1] 55sum(1:10) # Good commenting style[1] 55- Also, use commented lines of - and = to break up your file into easily readable sub-sections.
# Read data ----------------# Plot data ----------------To learn more read Hadley Wickham's Style guide.
Objects in R
Data structures are the ways of arranging data.
- You can create objects, using the left pointing arrow <-
Objects in R
Data structures are the ways of arranging data.
- You can create objects, using the left pointing arrow <-
Functions tell R to do something.
A function may be applied to an object.
Result of applying a function is usually an object too.
All function calls need to be followed by parentheses.
a <- 1:20 # data structuresum(a) # sum is a function applied on a[1] 210help.start() # Some functions work on their own.Getting help with functions and features
- R has inbuilt help facility
Method 1
help(rnorm)- For a feature specified by special characters such as
for,if,[[
help("[[")- Search the help files for a word or phrase.
help.search(‘weighted mean’)Method 2
?rnorm??rnormData structures
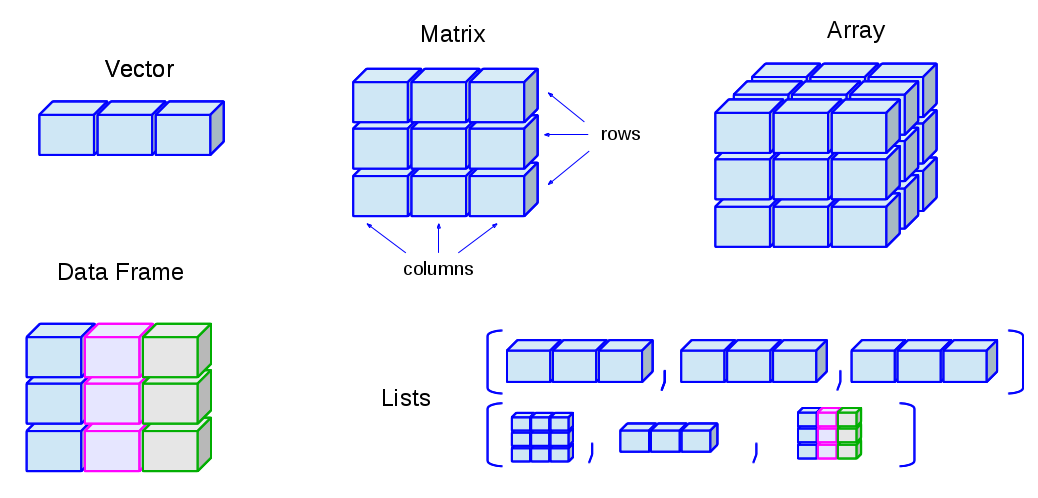
Data structures differ in terms of,
Type of data they can hold
How they are created
Structural complexity
Notation to identify and access individual elements
Image Credit: venus.ifca.unican.es
1. Vectors
Vectors
Vectors are one-dimensional arrays that can hold numeric data, character data, or logical data.
The function
cis used to form vectors.cstands for concatenate.Data in a vector must only be one type or mode (numeric, character, or logical). You can’t mix modes in the same vector.
Vector assignment
vector_name <- c(element1, element2, element3) # syntaxx <- c(5, 6, 3, 1 , 100) # exampleassignment operator ('<-'), '=' can be used as an alternative.
c()function is used to create vector.
Your turn
What will be the output of the following code?
x <- c(5, 6, 3, 1 , 100) xy <- c(x, 500, 600)y01:30
Types and tests with vectors
first_vec <- c(10, 20, 50, 70)second_vec <- c("Jan", "Feb", "March", "April")third_vec <- c(TRUE, FALSE, TRUE, TRUE)fourth_vec <- c(10L, 20L, 50L, 70L)To check if it is a
- vector:
is.vector()
is.vector(first_vec)[1] TRUE- character vector:
is.character()
is.character(first_vec)[1] FALSE- double:
is.double()
is.double(first_vec)[1] TRUE- integer:
is.integer()
is.integer(first_vec)[1] FALSE- logical:
is.logical()
is.logical(first_vec)[1] FALSE- length
length(first_vec)[1] 4Compare
first_vec <- c(10, 20, 50, 70)andfourth_vec <- c(10L, 20L, 50L, 70L)
is.double(fourth_vec)[1] FALSEis.integer(fourth_vec)[1] TRUEMathematical operations
sum(first_vec)[1] 150mean(first_vec)[1] 37.5summary(first_vec) Min. 1st Qu. Median Mean 3rd Qu. Max. 10.0 17.5 35.0 37.5 55.0 70.0More about functions: week 3.
Coercion
Vectors must be homogeneous. When you attempt to combine different types they will be coerced to the most flexible type so that every element in the vector is of the same type.
Order from least to most flexible
logical --> integer --> double --> character
a <- c(3.1, 2L, 3, 4, "GPA") typeof(a)[1] "character"anew <- c(3.1, 2L, 3, 4)typeof(anew)[1] "double"Explicit coercion
Vectors can be explicitly coerced from one class to another using the as.* functions, if available. For example, as.character, as.numeric, as.integer, and as.logical.
vec1 <- c(TRUE, FALSE, TRUE, TRUE)typeof(vec1)[1] "logical"vec2 <- as.integer(vec1)typeof(vec2)[1] "integer"vec2[1] 1 0 1 1Your turn
Why does the below output NAs?
x <- c("a", "b", "c")as.numeric(x)[1] NA NA NA02:00
Explicit coercion (cont.)
x1 <- 1:3x2 <- c(10, 20, 30)combinedx1x2 <- c(x1, x2)combinedx1x2[1] 1 2 3 10 20 30typeof(x1)[1] "integer"typeof(x2)[1] "double"typeof(combinedx1x2)[1] "double"Explicit coercion (cont.)
x1 <- 1:3x2 <- c(10, 20, 30)combinedx1x2 <- c(x1, x2)combinedx1x2[1] 1 2 3 10 20 30class(x1)[1] "integer"class(x2)[1] "numeric"class(combinedx1x2)[1] "numeric"- If you combine a numeric vector and a character vector
y1 <- c(1, 2, 3)y2 <- c("a", "b", "c")c(y1, y2)[1] "1" "2" "3" "a" "b" "c"Name elements in a vector
You can name elements in a vector in different ways. We will learn two of them.
- When creating it
x1 <- c(a=1991, b=1992, c=1993)x1 a b c 1991 1992 1993- Modifying the names of an existing vector
x2 <- c(1, 5, 10)names(x2) <- c("a", "b", "b")x2 a b b 1 5 10Note that the names do not have to be unique.
To remove names of a vector
Method 1
unname(x1); x1[1] 1991 1992 1993 a b c 1991 1992 1993Method 2
names(x2) <- NULL; x2[1] 1 5 10Your turn
What will be the output of the following code?
v <- c(1, 2, 3)names(v) <- c("a")v01:30
Simplifying vector creation: :
- colon
:produce regular spaced ascending or descending sequences.
10:16[1] 10 11 12 13 14 15 16-0.5:7.5[1] -0.5 0.5 1.5 2.5 3.5 4.5 5.5 6.5 7.57.5: -0.5[1] 7.5 6.5 5.5 4.5 3.5 2.5 1.5 0.5 -0.5-0.5:7.3[1] -0.5 0.5 1.5 2.5 3.5 4.5 5.5 6.5 class(10:16)[1] "integer"class(-0.5:7.5)[1] "numeric"class(7.5:-0.5)[1] "numeric"class(-0.5:7.3)[1] "numeric"Simplifying vector creation: seq
- sequence:
seq(initial_value, final_value, increment)
seq(0.5, 8)[1] 0.5 1.5 2.5 3.5 4.5 5.5 6.5 7.5seq(1,11) [1] 1 2 3 4 5 6 7 8 9 10 11seq(1, 11, length.out=5)[1] 1.0 3.5 6.0 8.5 11.0seq(0, 11, by=2)[1] 0 2 4 6 8 10Simplifying vector creation: rep
- repeats
rep()
rep(9, 5)[1] 9 9 9 9 9rep(1:4, 2)[1] 1 2 3 4 1 2 3 4rep(1:4, each=2) # each element is repeated twice[1] 1 1 2 2 3 3 4 4rep(1:4, times=2) # whole sequence is repeated twice[1] 1 2 3 4 1 2 3 4Simplifying vector creation: rep
virus <- rep(c("delta", "gamma"), times=3)virus[1] "delta" "gamma" "delta" "gamma" "delta" "gamma"virus <- rep(c("delta", "gamma"), times=3, length.out=5)virus[1] "delta" "gamma" "delta" "gamma" "delta"Simplifying vector creation: rep (cont.)
Write the output of the following codes.
Your turn:
rep(1:4, each=2, times=3)rep(1:4, 1:4)rep(1:4, c(4, 1, 4, 2))05:00
Logical operators
<=less than or equal to>=greater than or equal to|or&and<less than>greater than==equal
c(1, 2, 3) == c(10, 20, 3)[1] FALSE FALSE TRUEc(1, 2, 3) != c(10, 20, 3)[1] TRUE TRUE FALSE1:5 > 3[1] FALSE FALSE FALSE TRUE TRUE1:5 < 3[1] TRUE TRUE FALSE FALSE FALSEOperators: %in% - in the set
a <- c(1, 2, 3)b <- c(1, 10, 3)a %in% b[1] TRUE FALSE TRUEx <- 1:10; y <- 1:3x; y [1] 1 2 3 4 5 6 7 8 9 10[1] 1 2 3x %in% y [1] TRUE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSEy %in% x[1] TRUE TRUE TRUEVector arithmetic
- operations are performed element by element.
c(10, 100, 100) + 2 # two is added to every element in the vector[1] 12 102 102Vector arithmetic
- operations are performed element by element.
c(10, 100, 100) + 2 # two is added to every element in the vector[1] 12 102 102- operations between two vectors
v1 <- c(1, 2, 3); v2 <- c(10, 100, 1000)v1 + v2[1] 11 102 1003Vector arithmetic
- operations are performed element by element.
c(10, 100, 100) + 2 # two is added to every element in the vector[1] 12 102 102- operations between two vectors
v1 <- c(1, 2, 3); v2 <- c(10, 100, 1000)v1 + v2[1] 11 102 1003- Add two vectors of unequal length (length of the longer is an integer multiple of the length of the shorter vector)
longvec <- seq(10, 100, length=10); shortvec <- c(1, 2, 3, 4, 5)longvec + shortvec [1] 11 22 33 44 55 61 72 83 94 105- Add two vectors of unequal length (length of the longer is not an integer multiple of the length of the shorter vector)
# gives a warning message when the length of the longer is not an integer multiple of the length of the shorter vector.svec <- c(1, 2, 3)longvec + svec [1] 11 22 33 41 52 63 71 82 93 101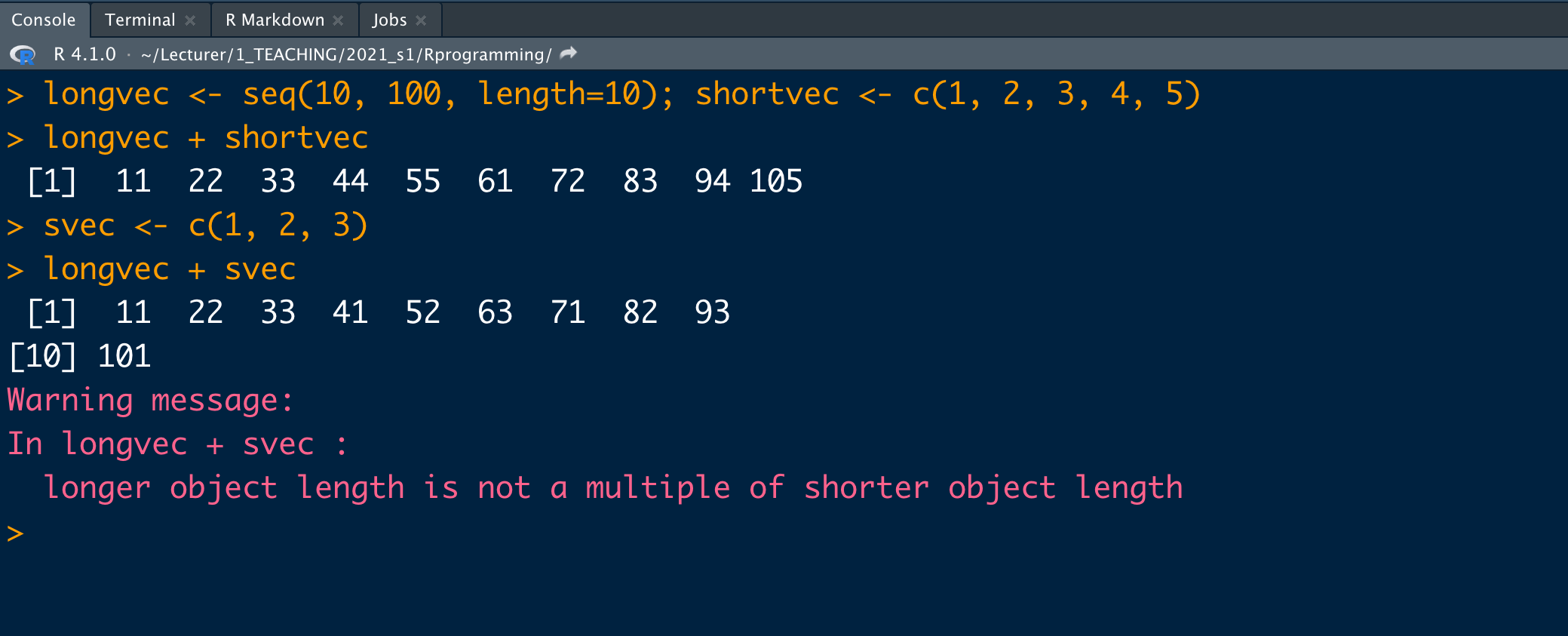
Your turn
What will be the output of the following code?
first <- c(1, 2, 3, 4); second <- c(10, 100)first * second02:30
Missing values
Use NA or NaN to place a missing value in a vector.
z <- c(10, 101, 2, 3, NA)is.na(z)[1] FALSE FALSE FALSE FALSE TRUEy <- c(10, 101, 2, 3, NaN)is.na(y)[1] FALSE FALSE FALSE FALSE TRUE